library(tidyverse)
library(stringr) # package for manipulating strings (part of tidyverse)
library(lubridate) # package for working with dates and times
#library(rvest) # package for reading and manipulating HTMLStrings & Dates
1 Introduction
Load packages:
If package not yet installed, then must install before you load. Install in “console” rather than .Rmd file:
- Generic syntax:
install.packages("package_name") - Install “tidyverse”:
install.packages("tidyverse")
Note: When we load package, name of package is not in quotes; but when we install package, name of package is in quotes:
install.packages("tidyverse")library(tidyverse)
Resources used to create this lecture:
- https://r4ds.had.co.nz/strings.html
- https://www.tutorialspoint.com/r/r_strings.htm
- https://swcarpentry.github.io/r-novice-inflammation/13-supp-data-structures/
- https://www.statmethods.net/input/datatypes.html
- https://www.stat.berkeley.edu/~s133/dates.html
1.1 Dataset we will use
We will use rtweet to pull Twitter data from the PAC-12 universities. We will use the university admissions Twitter handle if there is one, or the main Twitter handle for the university if there isn’t one:
- We wrote a short tutorial on using
rtweetin the Fall 2020 version of this class:
# library(rtweet)
#
# p12 <- c("uaadmissions", "FutureSunDevils", "caladmissions", "UCLAAdmission",
# "futurebuffs", "uoregon", "BeaverVIP", "USCAdmission",
# "engagestanford", "UtahAdmissions", "UW", "WSUPullman")
# p12_full_df <- search_tweets(paste0("from:", p12, collapse = " OR "), n = 500)
#
# saveRDS(p12_full_df, "p12_dataset.RDS")
# Load previously pulled Twitter data
# p12_full_df <- readRDS("p12_dataset.RDS")
p12_full_df <- readRDS(url("https://github.com/anyone-can-cook/rclass1/raw/master/data/twitter/p12_dataset.RDS", "rb"))
#glimpse(p12_full_df)
p12_df <- p12_full_df %>% select("user_id", "created_at", "screen_name", "text", "location")
head(p12_df)
#> # A tibble: 6 × 5
#> user_id created_at screen_name text location
#> <chr> <dttm> <chr> <chr> <chr>
#> 1 22080148 2020-04-25 22:37:18 WSUPullman "Big Dez is headed to Indy!… Pullman…
#> 2 22080148 2020-04-23 21:11:49 WSUPullman "Cougar Cheese. That's it. … Pullman…
#> 3 22080148 2020-04-21 04:00:00 WSUPullman "Darien McLaughlin '19, and… Pullman…
#> 4 22080148 2020-04-24 03:00:00 WSUPullman "6 houses, one pick. Cougs,… Pullman…
#> 5 22080148 2020-04-20 19:00:21 WSUPullman "Why did you choose to atte… Pullman…
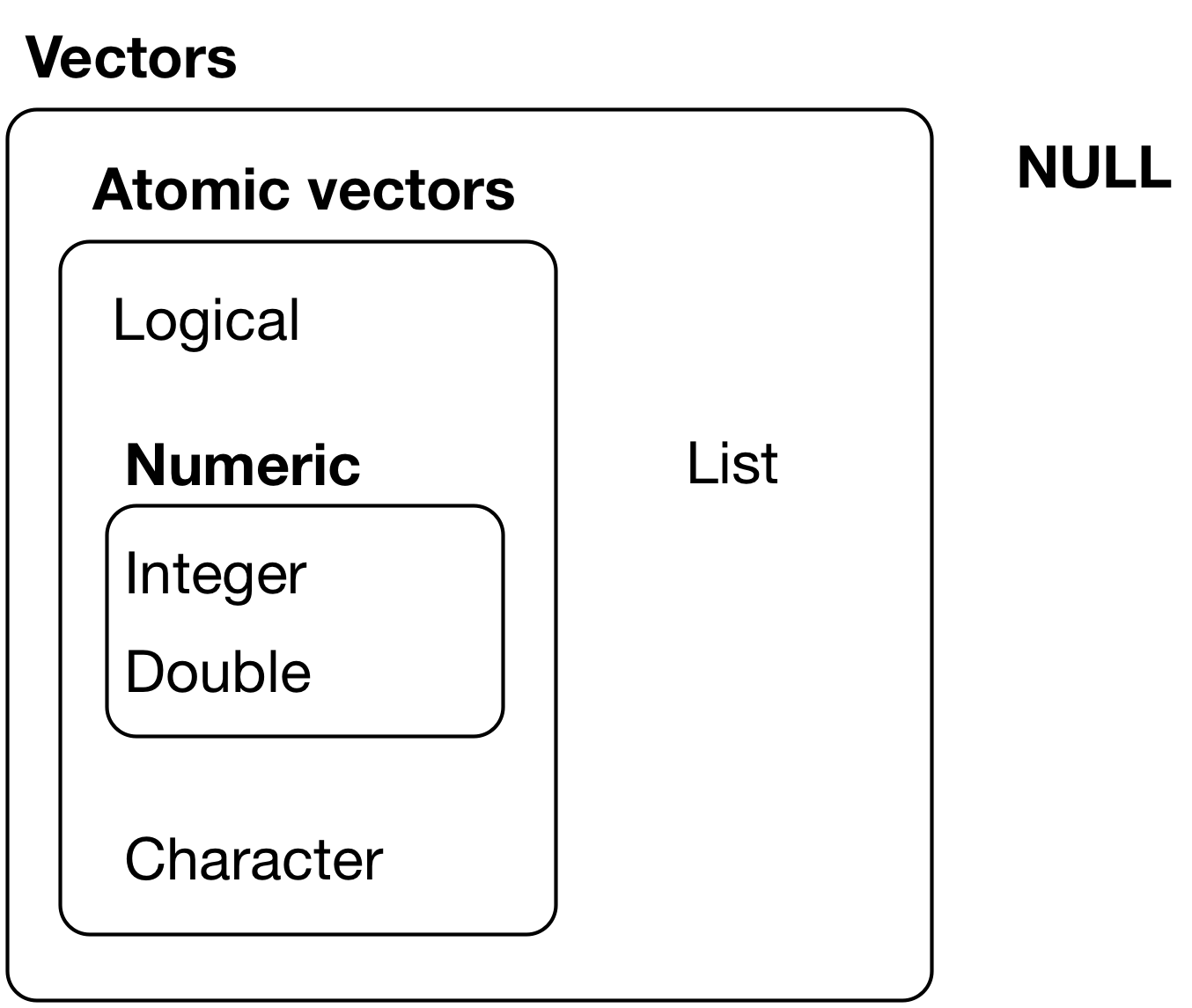
#> 6 22080148 2020-04-20 02:20:01 WSUPullman "Tell us one of your Bryan … Pullman…2 Review of Data structures and types
What is an object?
- Everything in R is an object
- We can classify objects based on their class and type
class(): What kind of object is it (high-level)?- The class of the object determines what kind of functions we can apply to it
typeof(): What is the object’s data type (low-level)?
- Objects may be combined to form data structures
Credit: R for Data Science
Basic data types:
- Logical (
TRUE,FALSE) - Numeric (e.g.,
5,2.5) - Integer (e.g.,
1L,4L, whereLtells R to store asintegertype) - Character (e.g.,
"R is fun")
Basic data structures:
Review from Intro to R and Attributes and Class lectures:
3 String basics
What are strings?
- String is a type of data in R
- You can create strings using either single quotes (
') or double quotes (")- Internally, R stores strings using double quotes
- The
class()andtypeof()a string ischaracter
Example: Creating string using single quotes
Notice how R stores strings using double quotes internally:
my_string <- 'This is a string'
my_string
#> [1] "This is a string"
Example: Creating string using double quotes
my_string <- "Strings can also contain numbers: 123"
my_string
#> [1] "Strings can also contain numbers: 123"
Example: Checking class and type of strings
class(my_string)
#> [1] "character"
typeof(my_string)
#> [1] "character"
Note: To include quotes as part of the string, we can either use the other type of quotes to surround the string (i.e., ' or ") or escape the quote using a backslash (\). We won’t be going in-depth into escaping characters for this class, but see appendix for more details if you are interested.
# Include quote by using the other type of quotes to surround the string
my_string <- "There's no issues with this string."
my_string
#> [1] "There's no issues with this string."
# Include quote of the same type by escaping it with a backslash
my_string <- 'There\'s no issues with this string.'
my_string
#> [1] "There's no issues with this string."# This would not work
my_string <- 'There's an issue with this string.'
my_string4 stringr package
“A consistent, simple and easy to use set of wrappers around the fantastic
stringipackage. All function and argument names (and positions) are consistent, all functions deal withNA’s and zero length vectors in the same way, and the output from one function is easy to feed into the input of another.”
Credit: stringrR documentation
The stringr package:
- The
stringrpackage is based off thestringipackage and is part of Tidyverse stringrcontains functions to work with strings- For many functions in the
stringrpackage, there are equivalent “base R” functions - But
stringrfunctions all follow the same rules, while rules often differ across different “base R” string functions, so we will focus exclusively onstringrfunctions - Most
stringrfunctions start withstr_(e.g.,str_length)
4.1 str_length()
The str_length() function:
?str_length
# SYNTAX
str_length(string)- Function: Find string length
- Arguments:
string: Character vector (or vector coercible to character)
- Note that
str_length()calculates the length of a string, whereas thelength()function (which is not part ofstringrpackage) calculates the number of elements in an object
Example: Using str_length() on string
str_length("cats")
#> [1] 4Compare to length(), which treats the string as a single object:
length("cats")
#> [1] 1Example: Using str_length() on a character vector
str_length(c("cats", "in", "hat"))
#> [1] 4 2 3Compare to length(), which finds the number of elements in the vector:
length(c("cats", "in", "hat"))
#> [1] 3
Example: Using str_length() on other vectors coercible to character
Logical vectors can be coerced to character vectors:
str_length(c(TRUE, FALSE))
#> [1] 4 5Numeric vectors can be coerced to character vectors:
str_length(c(1, 2.5, 3000))
#> [1] 1 3 4Integer vectors can be coerced to character vectors:
str_length(c(2L, 100L))
#> [1] 1 3
Example: Using str_length() on dataframe column
Recall that the columns in a dataframe are just vectors, so we can use str_length() as long as the vector is coercible to character type. Let’s look at the screen_name column from the p12_df:
# `p12_df` is a dataframe object
str(p12_df)
#> tibble [328 × 5] (S3: tbl_df/tbl/data.frame)
#> $ user_id : chr [1:328] "22080148" "22080148" "22080148" "22080148" ...
#> $ created_at : POSIXct[1:328], format: "2020-04-25 22:37:18" "2020-04-23 21:11:49" ...
#> $ screen_name: chr [1:328] "WSUPullman" "WSUPullman" "WSUPullman" "WSUPullman" ...
#> $ text : chr [1:328] "Big Dez is headed to Indy!\n\n#GoCougs | #NFLDraft2020 | @dadpat7 | @Colts | #NFLCougs https://t.co/NdGsvXnij7" "Cougar Cheese. That's it. That's the tweet. 🧀#WSU #GoCougs https://t.co/0OWGvQlRZs" "Darien McLaughlin '19, and her dog, Yuki, went on a #Pullman distance walk this weekend. We will let you judge "| __truncated__ "6 houses, one pick. Cougs, which one you got? Reply ⬇️ #WSU #CougsContain #GoCougs https://t.co/lNDx7r71b2" ...
#> $ location : chr [1:328] "Pullman, Washington USA" "Pullman, Washington USA" "Pullman, Washington USA" "Pullman, Washington USA" ...
# `screen_name` column is a character vector
str(p12_df$screen_name)
#> chr [1:328] "WSUPullman" "WSUPullman" "WSUPullman" "WSUPullman" ...
[Base R method] Use str_length() to calculate the length of each screen_name:
# Let's focus on just the unique screen names
unique(p12_df$screen_name)
#> [1] "WSUPullman" "CalAdmissions" "UW" "USCAdmission"
#> [5] "uoregon" "FutureSunDevils" "UCLAAdmission" "UtahAdmissions"
#> [9] "futurebuffs" "uaadmissions" "BeaverVIP"
str_length(unique(p12_df$screen_name))
#> [1] 10 13 2 12 7 15 13 14 11 12 9
[Tidyverse method] Use str_length() to calculate the length of each screen_name:
# Let's focus on just the unique screen names
p12_df %>% select(screen_name) %>% unique()
#> # A tibble: 11 × 1
#> screen_name
#> <chr>
#> 1 WSUPullman
#> 2 CalAdmissions
#> 3 UW
#> 4 USCAdmission
#> 5 uoregon
#> 6 FutureSunDevils
#> 7 UCLAAdmission
#> 8 UtahAdmissions
#> 9 futurebuffs
#> 10 uaadmissions
#> 11 BeaverVIP
#p12_df %>% select(screen_name) %>% unique() %>% str_length()Notice that the above line does not work as expected because we passed in a dataframe to str_length() and it is trying to coerce that to character:
class(p12_df %>% select(screen_name) %>% unique())
#> [1] "tbl_df" "tbl" "data.frame"An alternative way is to add a column to the dataframe that contains the result of applying str_length() to the screen_name vector:
p12_df %>% select(screen_name) %>% unique() %>%
mutate(screen_name_len = str_length(screen_name))
#> # A tibble: 11 × 2
#> screen_name screen_name_len
#> <chr> <int>
#> 1 WSUPullman 10
#> 2 CalAdmissions 13
#> 3 UW 2
#> 4 USCAdmission 12
#> 5 uoregon 7
#> 6 FutureSunDevils 15
#> 7 UCLAAdmission 13
#> 8 UtahAdmissions 14
#> 9 futurebuffs 11
#> 10 uaadmissions 12
#> 11 BeaverVIP 94.2 str_c()
The str_c() function:
?str_c
# SYNTAX AND DEFAULT VALUES
str_c(..., sep = "", collapse = NULL)- Function: Concatenate strings between vectors (element-wise)
- Arguments:
- The input is one or more character vectors (or vectors coercible to character)
- Zero length arguments are removed
- Scalar inputs (vectors of length 1) are recycled to the common length of vector inputs
sep: String to insert between input vectorscollapse: Optional string used to combine input vectors into single string
- The input is one or more character vectors (or vectors coercible to character)
Example: Using str_c() on one vector
Since we only provided one input vector, it has nothing to concatenate with, so str_c() will just return the same vector:
str_c(c("a", "b", "c"))
#> [1] "a" "b" "c"Note that specifying the sep argument will also not have any effect because we only have one input vector, and sep is the separator between multiple vectors:
str_c(c("a", "b", "c"), sep = "~")
#> [1] "a" "b" "c"
# Check length: Output is the original vector of 3 elements
str_c(c("a", "b", "c")) %>% length()
#> [1] 3As seen above, str_c() returns a vector by default (because the default value for the collapse argument is NULL). But we can specify a string for collapse in order to collapse the elements of the output vector into a single string:
str_c(c("a", "b", "c"), collapse = "|")
#> [1] "a|b|c"
# Check length: Output vector of length 3 is collapsed into a single string
str_c(c("a", "b", "c"), collapse = "|") %>% length()
#> [1] 1
# Check str_length: This gives the length of the collapsed string, which is 5 characters long
str_c(c("a", "b", "c"), collapse = "|") %>% str_length()
#> [1] 5
Example: Using str_c() on more than one vector
When we provide multiple input vectors, we can see that the vectors get concatenated element-wise (i.e., 1st element from each vector are concatenated, 2nd element from each vector are concatenated, etc):
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"))
#> [1] "ax!" "by?" "cz;"The default separator for each element-wise concatenation is an empty string (""), but we can customize that by specifying the sep argument:
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"), sep = "~")
#> [1] "a~x~!" "b~y~?" "c~z~;"
# Check length: Output vector is same length as input vectors
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"), sep = "~") %>% length()
#> [1] 3Again, we can specify the collapse argument in order to collapse the elements of the output vector into a single string:
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"), collapse = "|")
#> [1] "ax!|by?|cz;"
# Check length: Output vector of length 3 is collapsed into a single string
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"), collapse = "|") %>% length()
#> [1] 1
# Specifying both `sep` and `collapse`
str_c(c("a", "b", "c"), c("x", "y", "z"), c("!", "?", ";"), sep = "~", collapse = "|")
#> [1] "a~x~!|b~y~?|c~z~;"
Example: Using str_c() on “strings”
What do we mean by “strings”?
- Informally, We can think of a “string” as being a character vector with
length()equal to 1 (i.e., one element). - Another way to think of it, a “string” is anything you put in between quotes”.
- Loosely, we can also think of individual elements within a character vector as strings
Below, passing 3 strings into str_c() is like passing in 3 vectors of size 1 each.
- Remember that vectors are concatenated element-wise, so these strings will be joined like this:
str_c("a", "b", "c")
#> [1] "abc"
# Again, we can think of strings as being character vectors of size 1
str_c(c("a"), c("b"), c("c"))
#> [1] "abc"We can use sep to specify how the elements are separated:
str_c("a", "b", "c", sep = "~")
#> [1] "a~b~c"Since we only have 1 element in each vector, the output from str_c() is a vector of length 1. Thus, collapse will not be useful here since it works to collapse multiple elements in the output vector into a single string:
str_c("a", "b", "c", collapse = "|")
#> [1] "abc"
Example: Using str_c() on types other than character
When we provide a non-character vector (such as a numeric or logical vector), it will get coerced into a character vector:
str_c(c("a", "b", "c"), c(1, 2, 3), c(TRUE, FALSE, FALSE))
#> [1] "a1TRUE" "b2FALSE" "c3FALSE"
# Specifying both `sep` and `collapse`
str_c(c("a", "b", "c"), c(1, 2, 3), c(TRUE, FALSE, FALSE), sep = "~", collapse = "|")
#> [1] "a~1~TRUE|b~2~FALSE|c~3~FALSE"Note that we can also use any other single element input (other than string) that can be coerced to character:
str_c(TRUE, 1.5, 2L, "X")
#> [1] "TRUE1.52X"
Example: Using str_c() on vectors of different lengths
When trying to join vectors of different length, you will run into an error.
This is because new recycling rules are applied to the
stringr()package to be consistent with other tidyverse recycling rules (see Wickham blog 12/05/2022)[https://www.tidyverse.org/blog/2022/12/stringr-1-5-0/#recycling-rules]In practice this makes sense when working with data frames and tidyverse because columns (variables) in a data frame must be the same length.
str_c("#", c("a", "b", "c", "d"), c(1, 2, 3), c(TRUE, FALSE))
# Specifying both `sep` and `collapse`
str_c("#", c("a", "b", "c", "d"), c(1, 2, 3), c(TRUE, FALSE), sep = "~", collapse = "|")
Example: Using str_c() on dataframe columns
Let’s combine the user_id and screen_name columns from p12_df. We’ll focus on unique Twitter handles:
p12_unique_df <- p12_df %>% select(user_id, screen_name) %>% unique()
p12_unique_df
#> # A tibble: 11 × 2
#> user_id screen_name
#> <chr> <chr>
#> 1 22080148 WSUPullman
#> 2 15988549 CalAdmissions
#> 3 27103822 UW
#> 4 198643896 USCAdmission
#> 5 40940457 uoregon
#> 6 325014504 FutureSunDevils
#> 7 2938776590 UCLAAdmission
#> 8 4922145709 UtahAdmissions
#> 9 45879674 futurebuffs
#> 10 44733626 uaadmissions
#> 11 403743606 BeaverVIP
[Base R method] Use str_c() to combine user_id and screen_name:
str_c(p12_unique_df$user_id, "=", p12_unique_df$screen_name, sep = " ", collapse = ", ")
#> [1] "22080148 = WSUPullman, 15988549 = CalAdmissions, 27103822 = UW, 198643896 = USCAdmission, 40940457 = uoregon, 325014504 = FutureSunDevils, 2938776590 = UCLAAdmission, 4922145709 = UtahAdmissions, 45879674 = futurebuffs, 44733626 = uaadmissions, 403743606 = BeaverVIP"
str_c(p12_unique_df$user_id, "=", p12_unique_df$screen_name, sep = " ") # without collapsing to one element
#> [1] "22080148 = WSUPullman" "15988549 = CalAdmissions"
#> [3] "27103822 = UW" "198643896 = USCAdmission"
#> [5] "40940457 = uoregon" "325014504 = FutureSunDevils"
#> [7] "2938776590 = UCLAAdmission" "4922145709 = UtahAdmissions"
#> [9] "45879674 = futurebuffs" "44733626 = uaadmissions"
#> [11] "403743606 = BeaverVIP"
[Tidyverse method] Use str_c() to combine user_id and screen_name:
p12_unique_df %>% mutate(twitter_handle = str_c(user_id,screen_name))
#> # A tibble: 11 × 3
#> user_id screen_name twitter_handle
#> <chr> <chr> <chr>
#> 1 22080148 WSUPullman 22080148WSUPullman
#> 2 15988549 CalAdmissions 15988549CalAdmissions
#> 3 27103822 UW 27103822UW
#> 4 198643896 USCAdmission 198643896USCAdmission
#> 5 40940457 uoregon 40940457uoregon
#> 6 325014504 FutureSunDevils 325014504FutureSunDevils
#> 7 2938776590 UCLAAdmission 2938776590UCLAAdmission
#> 8 4922145709 UtahAdmissions 4922145709UtahAdmissions
#> 9 45879674 futurebuffs 45879674futurebuffs
#> 10 44733626 uaadmissions 44733626uaadmissions
#> 11 403743606 BeaverVIP 403743606BeaverVIP
p12_unique_df %>% mutate(twitter_handle = str_c("User #", user_id, " is @", screen_name))
#> # A tibble: 11 × 3
#> user_id screen_name twitter_handle
#> <chr> <chr> <chr>
#> 1 22080148 WSUPullman User #22080148 is @WSUPullman
#> 2 15988549 CalAdmissions User #15988549 is @CalAdmissions
#> 3 27103822 UW User #27103822 is @UW
#> 4 198643896 USCAdmission User #198643896 is @USCAdmission
#> 5 40940457 uoregon User #40940457 is @uoregon
#> 6 325014504 FutureSunDevils User #325014504 is @FutureSunDevils
#> 7 2938776590 UCLAAdmission User #2938776590 is @UCLAAdmission
#> 8 4922145709 UtahAdmissions User #4922145709 is @UtahAdmissions
#> 9 45879674 futurebuffs User #45879674 is @futurebuffs
#> 10 44733626 uaadmissions User #44733626 is @uaadmissions
#> 11 403743606 BeaverVIP User #403743606 is @BeaverVIP4.3 str_sub()
The str_sub() function:
?str_sub
# SYNTAX AND DEFAULT VALUES
str_sub(string, start = 1L, end = -1L)
str_sub(string, start = 1L, end = -1L, omit_na = FALSE) <- value- Function: Subset strings
- Arguments:
string: Character vector (or vector coercible to character)start: Position of first character to be included in substring (default:1)end: Position of last character to be included in substring (default:-1)- Negative index means counting backwards from the end of the string
- If an element in the vector is shorter than the specified
end, it will just include all the available characters that it does have
omit_na: IfTRUE, missing values in any of the arguments provided will result in an unchanged input
- When
str_sub()is used in the assignment form, you can replace the subsetted part of the string with avalueof your choice- If an element in the vector is too short to meet the subset specification, the replacement
valuewill be concatenated to the end of that element - Note that this modifies your input vector directly, so you must have the vector saved to a variable (see example below)
- If an element in the vector is too short to meet the subset specification, the replacement
Example: Using str_sub() to subset strings
If no start and end positions are specified, str_sub() will by default return the entire (original) string:
str_sub(string = c("abcdefg", 123, TRUE))
#> [1] "abcdefg" "123" "TRUE"Note that if an element is shorter than the specified end (i.e., 123 in the example below), it will just include all the available characters that it does have:
str_sub(string = c("abcdefg", 123, TRUE), start = 2, end = 4)
#> [1] "bcd" "23" "RUE"Remember we can also use negative index to count the position starting from the back:
str_sub(c("abcdefg", 123, TRUE), start = 2, end = -2)
#> [1] "bcdef" "2" "RU"
Example: Using str_sub() to replace strings
If no start and end positions are specified, str_sub() will by default return the original string, so the entire string would be replaced:
v <- c("A", "AB", "ABC", "ABCD", "ABCDE")
str_sub(v, start = 1,end =-1)
#> [1] "A" "AB" "ABC" "ABCD" "ABCDE"
str_sub(v, start = 1,end =-1) <- "*"
v
#> [1] "*" "*" "*" "*" "*"If an element in the vector is too short to meet the subset specification, the replacement value will be concatenated to the end of that element:
v <- c("A", "AB", "ABC", "ABCD", "ABCDE")
v
#> [1] "A" "AB" "ABC" "ABCD" "ABCDE"
str_sub(v, start = 2, end = 3)
#> [1] "" "B" "BC" "BC" "BC"
str_sub(v, start = 2, end = 3) <- "*"
v
#> [1] "A*" "A*" "A*" "A*D" "A*DE"Note that because the replacement form of str_sub() modifies the input vector directly, we need to save it in a variable first. Directly passing in the vector to str_sub() would give us an error:
# Does not work
str_sub(c("A", "AB", "ABC", "ABCD", "ABCDE")) <- "*"
Example: Using str_sub() on dataframe column
We can use as.character() to turn the created_at value to a string, then use str_sub() to extract out various date/time components from the string:
#str(p12_df %>% select(created_at))
typeof(p12_df$created_at)
#> [1] "double"
class(p12_df$created_at)
#> [1] "POSIXct" "POSIXt"
head(p12_df$created_at)
#> [1] "2020-04-25 22:37:18 UTC" "2020-04-23 21:11:49 UTC"
#> [3] "2020-04-21 04:00:00 UTC" "2020-04-24 03:00:00 UTC"
#> [5] "2020-04-20 19:00:21 UTC" "2020-04-20 02:20:01 UTC"
p12_datetime_df <- p12_df %>% select(created_at) %>%
mutate(
dt_chr = as.character(created_at), #convert to character
date_chr = str_sub(dt_chr, 1, 10), #subset values in position 1 and 10 to grab the date
yr_chr = str_sub(dt_chr, 1, 4), #subset values in position 1 and 4 to grab the year
mth_chr = str_sub(dt_chr, 6, 7),
day_chr = str_sub(dt_chr, 9, 10),
hr_chr = str_sub(dt_chr, -8, -7),
min_chr = str_sub(dt_chr, -5, -4),
sec_chr = str_sub(dt_chr, -2, -1)
)
p12_datetime_df
#> # A tibble: 328 × 9
#> created_at dt_chr date_chr yr_chr mth_chr day_chr hr_chr min_chr
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2020-04-25 22:37:18 2020-04-2… 2020-04… 2020 04 25 22 37
#> 2 2020-04-23 21:11:49 2020-04-2… 2020-04… 2020 04 23 21 11
#> 3 2020-04-21 04:00:00 2020-04-2… 2020-04… 2020 04 21 04 00
#> 4 2020-04-24 03:00:00 2020-04-2… 2020-04… 2020 04 24 03 00
#> 5 2020-04-20 19:00:21 2020-04-2… 2020-04… 2020 04 20 19 00
#> 6 2020-04-20 02:20:01 2020-04-2… 2020-04… 2020 04 20 02 20
#> 7 2020-04-22 04:00:00 2020-04-2… 2020-04… 2020 04 22 04 00
#> 8 2020-04-25 17:00:00 2020-04-2… 2020-04… 2020 04 25 17 00
#> 9 2020-04-21 15:13:06 2020-04-2… 2020-04… 2020 04 21 15 13
#> 10 2020-04-21 17:52:47 2020-04-2… 2020-04… 2020 04 21 17 52
#> # ℹ 318 more rows
#> # ℹ 1 more variable: sec_chr <chr>4.4 Other stringr functions
Other useful stringr functions:
str_to_upper(): Turn strings to uppercasestr_to_lower(): Turn strings to lowercasestr_sort(): Sort a character vectorstr_trim(): Trim whitespace from strings (including\n,\t, etc.)str_pad(): Pad strings with specified character
Example: Using str_to_upper() to turn strings to uppercase
Turn column names of p12_df to uppercase:
# Column names are originally lowercase
names(p12_df)
#> [1] "user_id" "created_at" "screen_name" "text" "location"
# Turn column names to uppercase
names(p12_df) <- str_to_upper(names(p12_df))
names(p12_df)
#> [1] "USER_ID" "CREATED_AT" "SCREEN_NAME" "TEXT" "LOCATION"
Example: Using str_to_lower() to turn strings to lowercase
Turn column names of p12_df to lowercase:
# Column names are originally uppercase
names(p12_df)
#> [1] "USER_ID" "CREATED_AT" "SCREEN_NAME" "TEXT" "LOCATION"
# Turn column names to lowercase
names(p12_df) <- str_to_lower(names(p12_df))
names(p12_df)
#> [1] "user_id" "created_at" "screen_name" "text" "location"
Example: Using str_sort() to sort character vector
Sort the vector of p12_df column names:
# Before sort
names(p12_df)
#> [1] "user_id" "created_at" "screen_name" "text" "location"
# Sort alphabetically (default)
str_sort(names(p12_df))
#> [1] "created_at" "location" "screen_name" "text" "user_id"
# Sort reverse alphabetically
str_sort(names(p12_df), decreasing = TRUE)
#> [1] "user_id" "text" "screen_name" "location" "created_at"
Example: Using str_trim() to trim whitespace from string
# Trim whitespace from both left and right sides (default)
str_trim(c("\nABC ", " XYZ\t"))
#> [1] "ABC" "XYZ"
# Trim whitespace from left side
str_trim(c("\nABC ", " XYZ\t"), side = "left")
#> [1] "ABC " "XYZ\t"
# Trim whitespace from right side
str_trim(c("\nABC ", " XYZ\t"), side = "right")
#> [1] "\nABC" " XYZ"
Example: Using str_pad() to pad string with character
Let’s say we have a vector of zip codes that has lost all leading 0’s. We can use str_pad() to add that back in:
# Pad the left side of strings with "0" until width of 5 is reached
str_pad(c(95035, 90024, 5009, 5030), width = 5, side = "left", pad = "0")
#> [1] "95035" "90024" "05009" "05030"5 Dates and times
“Date-time data can be frustrating to work with in R. R commands for date-times are generally unintuitive and change depending on the type of date-time object being used. Moreover, the methods we use with date-times must be robust to time zones, leap days, daylight savings times, and other time related quirks, and R lacks these capabilities in some situations. Lubridate makes it easier to do the things R does with date-times and possible to do the things R does not.”
Credit: lubridatedocumentation
How are dates and times stored in R? (From Dates and Times in R)
- The
Dateclass is used for storing dates- “Internally,
Dateobjects are stored as the number of days since January 1, 1970, using negative numbers for earlier dates. Theas.numeric()function can be used to convert aDateobject to its internal form.”
- “Internally,
- POSIX classes can be used for storing date plus times
- “The
POSIXctclass stores date/time values as the number of seconds since January 1, 1970” - “The
POSIXltclass stores date/time values as a list of components (hour, min, sec, mon, etc.) making it easy to extract these parts”
- “The
- There is no native R class for storing only time
Why use date/time objects?
- Using date/time objects makes it easier to fetch or modify various date/time components (e.g., year, month, day, day of the week)
- Compared to if the date/time is just stored in a string, these components are not as readily accessible and need to be parsed
- You can perform certain arithmetics with date/time objects (e.g., find the “difference” between date/time points)
5.1 Creating date/time objects by parsing input
Functions that create date/time objects by parsing character or numeric input:
- Create
Dateobject:ymd(),ydm(),mdy(),myd(),dmy(), anddym()ystands for year,mstands for month,dstands for day- Select the function that represents the order in which your date input is formatted, and the function will be able to parse your input and create a
Dateobject
- Create
POSIXctobject:ymd_h(),ymd_hm(),ymd_hms(), etc.hstands for hour,mstands for minute,sstands for second- For any of the previous 6 date functions, you can append
h,hm, orhmsif you want to provide additional time information in order to create aPOSIXctobject - To force a
POSIXctobject without providing any time information, you can just provide a timezone (usingtz) to one of the date functions and it will assume midnight as the time - You can use
Sys.timezone()to get the timezone for your location
Example: Creating Date object from character or numeric input
The lubridate functions are flexible and can parse dates in various formats:
d <- mdy("1/1/2024")
d
#> [1] "2024-01-01"
d <- mdy("1-1-2024")
d
#> [1] "2024-01-01"
d <- mdy("Jan. 1, 2024")
d
#> [1] "2024-01-01"
d <- ymd(20240101)
d
#> [1] "2024-01-01"
Investigate the Date object:
class(d)
#> [1] "Date"
typeof(d)
#> [1] "double"
# Number of days since January 1, 1970
as.numeric(d)
#> [1] 19723
Example: Creating Date objects from dataframe column
Using the p12_datetime_df we created earlier, we can create Date objects from the date_chr column:
# Use `ymd()` to parse the string stored in the `date_chr` column
p12_datetime_df %>% select(created_at, dt_chr, date_chr) %>%
mutate(date_ymd = ymd(date_chr))
#> # A tibble: 328 × 4
#> created_at dt_chr date_chr date_ymd
#> <dttm> <chr> <chr> <date>
#> 1 2020-04-25 22:37:18 2020-04-25 22:37:18 2020-04-25 2020-04-25
#> 2 2020-04-23 21:11:49 2020-04-23 21:11:49 2020-04-23 2020-04-23
#> 3 2020-04-21 04:00:00 2020-04-21 04:00:00 2020-04-21 2020-04-21
#> 4 2020-04-24 03:00:00 2020-04-24 03:00:00 2020-04-24 2020-04-24
#> 5 2020-04-20 19:00:21 2020-04-20 19:00:21 2020-04-20 2020-04-20
#> 6 2020-04-20 02:20:01 2020-04-20 02:20:01 2020-04-20 2020-04-20
#> 7 2020-04-22 04:00:00 2020-04-22 04:00:00 2020-04-22 2020-04-22
#> 8 2020-04-25 17:00:00 2020-04-25 17:00:00 2020-04-25 2020-04-25
#> 9 2020-04-21 15:13:06 2020-04-21 15:13:06 2020-04-21 2020-04-21
#> 10 2020-04-21 17:52:47 2020-04-21 17:52:47 2020-04-21 2020-04-21
#> # ℹ 318 more rows
Example: Creating POSIXct object from character or numeric input
The lubridate functions are flexible and can parse AM/PM in various formats:
dt <- mdy_h("12/31/2023 11pm")
dt
#> [1] "2023-12-31 23:00:00 UTC"
dt <- mdy_hm("12/31/2023 11:59 pm")
dt
#> [1] "2023-12-31 23:59:00 UTC"
dt <- mdy_hms("12/31/2023 11:59:59 PM")
dt
#> [1] "2023-12-31 23:59:59 UTC"
dt <- ymd_hms(20231231235959)
dt
#> [1] "2023-12-31 23:59:59 UTC"
Investigate the POSIXct object:
class(dt)
#> [1] "POSIXct" "POSIXt"
typeof(dt)
#> [1] "double"
# Number of seconds since January 1, 1970
as.numeric(dt)
#> [1] 1704067199
We can also create a POSIXct object from a date function by providing a timezone. The time would default to midnight:
dt <- mdy("1/1/2024", tz = "UTC")
dt
#> [1] "2024-01-01 UTC"
# Number of seconds since January 1, 1970
as.numeric(dt) # Note that this is indeed 1 sec after the previous example
#> [1] 1704067200
Example: Creating POSIXct objects from dataframe column
Using the p12_datetime_df we created earlier, we can recreate the created_at column (class POSIXct) from the dt_chr column (class character):
# Use `ymd_hms()` to parse the string stored in the `dt_chr` column
p12_datetime_df %>% select(created_at, dt_chr) %>%
mutate(datetime_ymd_hms = ymd_hms(dt_chr))
#> # A tibble: 328 × 3
#> created_at dt_chr datetime_ymd_hms
#> <dttm> <chr> <dttm>
#> 1 2020-04-25 22:37:18 2020-04-25 22:37:18 2020-04-25 22:37:18
#> 2 2020-04-23 21:11:49 2020-04-23 21:11:49 2020-04-23 21:11:49
#> 3 2020-04-21 04:00:00 2020-04-21 04:00:00 2020-04-21 04:00:00
#> 4 2020-04-24 03:00:00 2020-04-24 03:00:00 2020-04-24 03:00:00
#> 5 2020-04-20 19:00:21 2020-04-20 19:00:21 2020-04-20 19:00:21
#> 6 2020-04-20 02:20:01 2020-04-20 02:20:01 2020-04-20 02:20:01
#> 7 2020-04-22 04:00:00 2020-04-22 04:00:00 2020-04-22 04:00:00
#> 8 2020-04-25 17:00:00 2020-04-25 17:00:00 2020-04-25 17:00:00
#> 9 2020-04-21 15:13:06 2020-04-21 15:13:06 2020-04-21 15:13:06
#> 10 2020-04-21 17:52:47 2020-04-21 17:52:47 2020-04-21 17:52:47
#> # ℹ 318 more rows5.2 Creating date/time objects from individual components
Functions that create date/time objects from various date/time components:
- Create
Dateobject:make_date()- Syntax and default values:
make_date(year = 1970L, month = 1L, day = 1L) - All inputs are coerced to integer
- Syntax and default values:
- Create
POSIXctobject:make_datetime()- Syntax and default values:
make_datetime(year = 1970L, month = 1L, day = 1L, hour = 0L, min = 0L, sec = 0, tz = "UTC") - Input values should be numeric
- Syntax and default values:
Example: Creating Date object from individual components
There are various ways to pass in the inputs to create the same Date object:
d <- make_date(2024, 1, 1)
d
#> [1] "2024-01-01"
# Characters can be coerced to integers
d <- make_date("2024", "01", "01")
d
#> [1] "2024-01-01"
# Remember that the default values for month and day would be 1L
d <- make_date(2024)
d
#> [1] "2024-01-01"
Example: Creating Date objects from dataframe columns
Using the p12_datetime_df we created earlier, we can create Date objects from the various date component columns:
# Use `make_date()` to create a `Date` object from the `yr_chr`, `mth_chr`, `day_chr` fields
p12_datetime_df %>% select(created_at, dt_chr, yr_chr, mth_chr, day_chr) %>%
mutate(date_make_date = make_date(year = yr_chr, month = mth_chr, day = day_chr))
#> # A tibble: 328 × 6
#> created_at dt_chr yr_chr mth_chr day_chr date_make_date
#> <dttm> <chr> <chr> <chr> <chr> <date>
#> 1 2020-04-25 22:37:18 2020-04-25 22:37:18 2020 04 25 2020-04-25
#> 2 2020-04-23 21:11:49 2020-04-23 21:11:49 2020 04 23 2020-04-23
#> 3 2020-04-21 04:00:00 2020-04-21 04:00:00 2020 04 21 2020-04-21
#> 4 2020-04-24 03:00:00 2020-04-24 03:00:00 2020 04 24 2020-04-24
#> 5 2020-04-20 19:00:21 2020-04-20 19:00:21 2020 04 20 2020-04-20
#> 6 2020-04-20 02:20:01 2020-04-20 02:20:01 2020 04 20 2020-04-20
#> 7 2020-04-22 04:00:00 2020-04-22 04:00:00 2020 04 22 2020-04-22
#> 8 2020-04-25 17:00:00 2020-04-25 17:00:00 2020 04 25 2020-04-25
#> 9 2020-04-21 15:13:06 2020-04-21 15:13:06 2020 04 21 2020-04-21
#> 10 2020-04-21 17:52:47 2020-04-21 17:52:47 2020 04 21 2020-04-21
#> # ℹ 318 more rows
Example: Creating POSIXct object from individual components
# Inputs should be numeric
d <- make_datetime(2023, 12, 31, 23, 59, 59)
d
#> [1] "2023-12-31 23:59:59 UTC"
Example: Creating POSIXct objects from dataframe columns
Using the p12_datetime_df we created earlier, we can recreate the created_at column (class POSIXct) from the various date and time component columns (class character):
# Use `make_datetime()` to create a `POSIXct` object from the `yr_chr`, `mth_chr`, `day_chr`, `hr_chr`, `min_chr`, `sec_chr` fields
# Convert inputs to integers first
p12_datetime_df %>%
mutate(datetime_make_datetime = make_datetime(
as.integer(yr_chr), as.integer(mth_chr), as.integer(day_chr),
as.integer(hr_chr), as.integer(min_chr), as.integer(sec_chr)
)) %>%
select(datetime_make_datetime, yr_chr, mth_chr, day_chr, hr_chr, min_chr, sec_chr)
#> # A tibble: 328 × 7
#> datetime_make_datetime yr_chr mth_chr day_chr hr_chr min_chr sec_chr
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2020-04-25 22:37:18 2020 04 25 22 37 18
#> 2 2020-04-23 21:11:49 2020 04 23 21 11 49
#> 3 2020-04-21 04:00:00 2020 04 21 04 00 00
#> 4 2020-04-24 03:00:00 2020 04 24 03 00 00
#> 5 2020-04-20 19:00:21 2020 04 20 19 00 21
#> 6 2020-04-20 02:20:01 2020 04 20 02 20 01
#> 7 2020-04-22 04:00:00 2020 04 22 04 00 00
#> 8 2020-04-25 17:00:00 2020 04 25 17 00 00
#> 9 2020-04-21 15:13:06 2020 04 21 15 13 06
#> 10 2020-04-21 17:52:47 2020 04 21 17 52 47
#> # ℹ 318 more rows5.3 Date/time object components
Storing data using date/time objects makes it easier to get and set the various date/time components.
- Basic accessor functions:
date(): Date componentyear(): Yearmonth(): Monthday(): Dayhour(): Hourminute(): Minutesecond(): Secondweek(): Week of the yearwday(): Day of the week (1for Sunday to7for Saturday)am(): Is it in the am? (returnsTRUEorFALSE)pm(): Is it in the pm? (returnsTRUEorFALSE)
- To get a date/time component, you can simply pass a date/time object to the function
- Syntax:
accessor_function(<date/time_object>)
- Syntax:
- To set a date/time component, you can assign into the accessor function to change the component
- Syntax:
accessor_function(<date/time_object>) <- "new_component" - Note that
am()andpm()can’t be set. Modify the time components instead.
- Syntax:
Example: Getting date/time components
# Create datetime for New Year's Eve
dt <- make_datetime(2023, 12, 31, 23, 59, 59)
dt
#> [1] "2023-12-31 23:59:59 UTC"
dt %>% class()
#> [1] "POSIXct" "POSIXt"
# Get date
date(dt)
#> [1] "2023-12-31"
# Get hour
hour(dt)
#> [1] 23
# Is it pm?
pm(dt)
#> [1] TRUE
# Day of the week (1 = Sunday)
wday(dt)
#> [1] 1
year(dt)
#> [1] 2023Example: Setting date/time components
# Create datetime for New Year's Eve
dt <- make_datetime(2023, 12, 31, 23, 59, 59)
dt
#> [1] "2023-12-31 23:59:59 UTC"
# Get week of year
week(dt)
#> [1] 53
# Set week of year (move back 1 week)
week(dt) <- week(dt) - 1
# Date now moved from New Year's Eve to Christmas Eve
dt
#> [1] "2023-12-24 23:59:59 UTC"
# Set day to Christmas Day
day(dt) <- 25
# Date now moved from Christmas Eve to Christmas Day
dt
#> [1] "2023-12-25 23:59:59 UTC"Example: Getting date/time components from dataframe column
Using the p12_datetime_df we created earlier, we can isolate the various date/time components from the POSIXct object in the created_at column:
# The extracted date/time components will be of numeric type
p12_datetime_df %>% select(created_at) %>%
mutate(
yr_num = year(created_at),
mth_num = month(created_at),
day_num = day(created_at),
hr_num = hour(created_at),
min_num = minute(created_at),
sec_num = second(created_at),
ampm = ifelse(am(created_at), 'AM', 'PM') # am()/pm() returns TRUE/FALSE
)
#> # A tibble: 328 × 8
#> created_at yr_num mth_num day_num hr_num min_num sec_num ampm
#> <dttm> <dbl> <dbl> <int> <int> <int> <dbl> <chr>
#> 1 2020-04-25 22:37:18 2020 4 25 22 37 18 PM
#> 2 2020-04-23 21:11:49 2020 4 23 21 11 49 PM
#> 3 2020-04-21 04:00:00 2020 4 21 4 0 0 AM
#> 4 2020-04-24 03:00:00 2020 4 24 3 0 0 AM
#> 5 2020-04-20 19:00:21 2020 4 20 19 0 21 PM
#> 6 2020-04-20 02:20:01 2020 4 20 2 20 1 AM
#> 7 2020-04-22 04:00:00 2020 4 22 4 0 0 AM
#> 8 2020-04-25 17:00:00 2020 4 25 17 0 0 PM
#> 9 2020-04-21 15:13:06 2020 4 21 15 13 6 PM
#> 10 2020-04-21 17:52:47 2020 4 21 17 52 47 PM
#> # ℹ 318 more rows5.4 Time spans
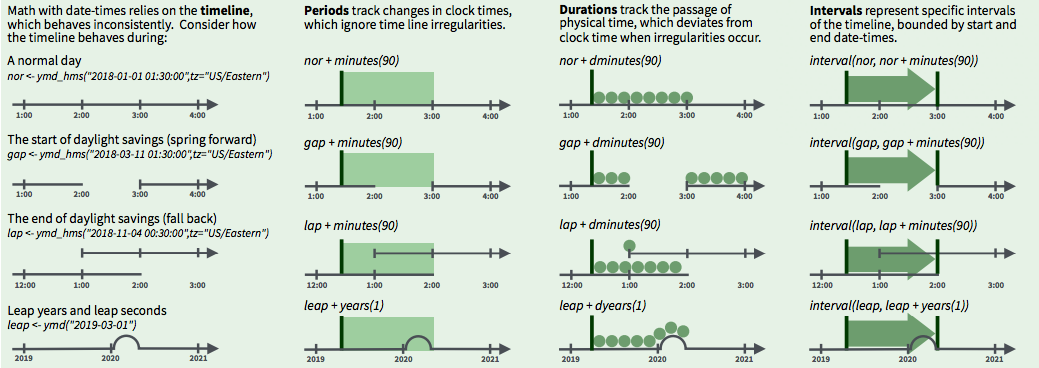
3 ways to represent time spans (From lubridate cheatsheet)
- Intervals represent specific intervals of the timeline, bounded by start and end date-times
- Example: People with birthdays between the interval October 23 to November 22 are Scorpios
- Periods track changes in clock times, which ignore time line irregularities
- Example: Daylight savings time ends at the beginning of November and we gain an hour - this extra hour is ignored when determining the period between October 23 to November 22
- Durations track the passage of physical time, which deviates from clock time when irregularities occur
- Example: Daylight savings time ends at the beginning of November and we gain an hour - this extra hour is added when determining the duration between October 23 to November 22
Time spans using lubridate
Using the lubridate package for time spans:
- Interval
- Create an interval using
interval()or%--%- Syntax:
interval(<date/time_object1>, <date/time_object2>)or<date/time_object1> %--% <date/time_object2>
- Syntax:
- Create an interval using
- Periods
- “Periods are time spans but don’t have a fixed length in seconds, instead they work with ‘human’ times, like days and months.” (From R for Data Science)
- Create periods using functions whose name is the time unit pluralized (e.g.,
years(),months(),weeks(),days(),hours(),minutes(),seconds())Example:
days(1)creates a period of 1 day - it does not matter if this day happened to have an extra hour due to daylight savings ending, since periods do not have a physical lengthdays(1) #> [1] "1d 0H 0M 0S"
- You can add and subtract periods
- You can also use
as.period()to get period of an interval
- Durations
- Durations keep track of the physical amount of time elapsed, so it is “stored as seconds, the only time unit with a consistent length” (From lubridate cheatsheet)
- Create durations using functions whose name is the time unit prefixed with a
d(e.g.,dyears(),dweeks(),ddays(),dhours(),dminutes(),dseconds())Example:
ddays(1)creates a duration of86400s, using the standard conversion of60seconds in an minute,60minutes in an hour, and24hours in a day:ddays(1) #> [1] "86400s (~1 days)"Notice that the output says this is equivalent to approximately
1day, since it acknowledges that not all days have24hours. In the case of daylight savings, one particular day may have25hours, so the duration of that day should be represented as:ddays(1) + dhours(1) #> [1] "90000s (~1.04 days)"
- You can add and subract durations
- You can also use
as.duration()to get duration of an interval
Example: Working with interval
# Use `Sys.timezone()` to get timezone for your location (time is midnight by default)
scorpio_start <- ymd("2023-10-23", tz = Sys.timezone())
scorpio_end <- ymd("2023-11-22", tz = Sys.timezone())
scorpio_start
#> [1] "2023-10-23 PDT"
# These datetime objects have class `POSIXct`
class(scorpio_start)
#> [1] "POSIXct" "POSIXt"
# Create interval for the datetimes
scorpio_interval <- scorpio_start %--% scorpio_end # or `interval(scorpio_start, scorpio_end)`
scorpio_interval <- interval(scorpio_start, scorpio_end)
scorpio_interval
#> [1] 2023-10-23 PDT--2023-11-22 PST
# The object has class `Interval`
class(scorpio_interval)
#> [1] "Interval"
#> attr(,"package")
#> [1] "lubridate"
as.numeric(scorpio_interval)
#> [1] 2595600Example: Working with period
If we use as.period() to get the period of scorpio_interval, we see that it is a period of 30 days. We do not worry about the extra 1 hour gained due to daylight savings ending:
# Period is 30 days
scorpio_period <- as.period(scorpio_interval)
scorpio_period
#> [1] "30d 0H 0M 0S"
# The object has class `Period`
class(scorpio_period)
#> [1] "Period"
#> attr(,"package")
#> [1] "lubridate"
Because periods work with “human” times like days, it is more intuitive. For example, if we add a period of 30 days to the scorpio_start datetime object, we get the expected end datetime that is 30 days later:
# Start datetime for Scorpio birthdays (time is midnight)
scorpio_start
#> [1] "2023-10-23 PDT"
# After adding 30 day period, we get the expected end datetime (time is midnight)
scorpio_start + days(30)
#> [1] "2023-11-22 PST"Example: Working with duration
If we use as.duration() to get the duration of scorpio_interval, we see that it is a duration of 2595600 seconds. It takes into account the extra 1 hour gained due to daylight savings ending:
# Duration is 2595600 seconds, which is equivalent to 30 24-hr days + 1 additional hour
scorpio_duration <- as.duration(scorpio_interval)
scorpio_duration
#> [1] "2595600s (~4.29 weeks)"
# The object has class `Duration`
class(scorpio_duration)
#> [1] "Duration"
#> attr(,"package")
#> [1] "lubridate"
# Using the standard 60s/min, 60min/hr, 24hr/day conversion,
# confirm duration is slightly more than 30 "standard" (ie. 24-hr) days
2595600 / (60 * 60 * 24)
#> [1] 30.04167
# Specifically, it is 30 days + 1 hour, if we define a day to have 24 hours
seconds_to_period(scorpio_duration)
#> [1] "30d 1H 0M 0S"
Because durations work with physical time, when we add a duration of 30 days to the scorpio_start datetime object, we do not get the end datetime we’d expect:
# Start datetime for Scorpio birthdays (time is midnight)
scorpio_start
#> [1] "2023-10-23 PDT"
# After adding 30 day duration, we do not get the expected end datetime
# `ddays(30)` adds the number of seconds in 30 standard 24-hr days, but one of the days has 25 hours
scorpio_start + ddays(30)
#> [1] "2023-11-21 23:00:00 PST"
# We need to add the additional 1 hour of physical time that elapsed during this time span
scorpio_start + ddays(30) + dhours(1)
#> [1] "2023-11-22 PST"6 Appendix
6.1 Special Characters
“A sequence in a string that starts with a
\is called an escape sequence and allows us to include special characters in our strings.”
Credit: Escape sequences from DataCamp
Special characters are characters that will not be interpreted literally.
Common special characters:
\n: newline\t: tab\: used for escaping purposes\': literal single quote\": literal double quote\\: literal backslash
These characters followed by a backslash \ take on a new meaning. The n by itself is just an n. When you add a backslash to the \n you are escaping it and making it a special character where \n now represents a newline.
The writeLines() function:
?writeLines
# SYNTAX AND DEFAULT VALUES
writeLines(text, con = stdout(), sep = "\n", useBytes = FALSE)- “
writeLines()displays quotes and backslashes as they would be read, rather than as R stores them.” (From writeLines documentation) - When we include escape sequences in the string, it is helpful to use
writeLines()to see how the escaped string looks writeLines()will also output the string without showing the outer pair of double quotes that R uses to store it, so we only see the content of the string
Example: Escaping single quotes
my_string <- 'Escaping single quote \' within single quotes'
my_string
#> [1] "Escaping single quote ' within single quotes"Alternatively, we could’ve just created the string using double quotes:
my_string <- "Single quote ' within double quotes does not need escaping"
my_string
#> [1] "Single quote ' within double quotes does not need escaping"Using writeLines() shows us only the content of the string without the outer pair of double quotes that R uses to store strings:
writeLines(my_string)
#> Single quote ' within double quotes does not need escapingExample: Escaping double quotes
my_string <- "Escaping double quote \" within double quotes"
my_string
#> [1] "Escaping double quote \" within double quotes"Alternatively, we could’ve just created the string using single quotes:
my_string <- 'Double quote " within single quotes does not need escaping'
my_string
#> [1] "Double quote \" within single quotes does not need escaping"Notice how the backslash still showed up in the above output to escape our double quote from the outer pair of double quotes that R uses to store the string. This is no longer an issue if we use writeLines() to only show the string content:
writeLines(my_string)
#> Double quote " within single quotes does not need escapingExample: Escaping double quotes within double quotes
my_string <- "I called my mom and she said \"Echale ganas!\""
my_string
#> [1] "I called my mom and she said \"Echale ganas!\""Using writeLines() shows us only the content of the string without the backslashes:
writeLines(my_string)
#> I called my mom and she said "Echale ganas!"Example: Escaping backslashes
To include a literal backslash in the string, we need to escape the backslash with another backslash:
my_string <- "The executable is located in C:\\Program Files\\Git\\bin"
my_string
#> [1] "The executable is located in C:\\Program Files\\Git\\bin"Use writeLines() to see the escaped string:
writeLines(my_string)
#> The executable is located in C:\Program Files\Git\binExample: Other special characters
my_string <- "A\tB\nC\tD"
my_string
#> [1] "A\tB\nC\tD"Use writeLines() to see the escaped string:
writeLines(my_string)
#> A B
#> C DEscape special characters using Twitter data
Let’s take a look at some tweets from our PAC-12 universities.
- Let’s start by grabbing observations 1-3 from the
textcolumn.
#Twitter example of \n newline special characters
p12_df$text[1:3]
#> [1] "Big Dez is headed to Indy!\n\n#GoCougs | #NFLDraft2020 | @dadpat7 | @Colts | #NFLCougs https://t.co/NdGsvXnij7"
#> [2] "Cougar Cheese. That's it. That's the tweet. 🧀#WSU #GoCougs https://t.co/0OWGvQlRZs"
#> [3] "Darien McLaughlin '19, and her dog, Yuki, went on a #Pullman distance walk this weekend. We will let you judge who was leading the way.🚶♀️🐕\n\nTweet a pic of how you are social distancing w/ the hashtag #CougsContain & tag @WSUPullman #GoCougs https://t.co/EltXDy1tPt"- Using
writeLines()we can see the contents of the strings as they would be read, rather than as R stores them.
writeLines(p12_df$text[1:3])
#> Big Dez is headed to Indy!
#>
#> #GoCougs | #NFLDraft2020 | @dadpat7 | @Colts | #NFLCougs https://t.co/NdGsvXnij7
#> Cougar Cheese. That's it. That's the tweet. 🧀#WSU #GoCougs https://t.co/0OWGvQlRZs
#> Darien McLaughlin '19, and her dog, Yuki, went on a #Pullman distance walk this weekend. We will let you judge who was leading the way.🚶♀️🐕
#>
#> Tweet a pic of how you are social distancing w/ the hashtag #CougsContain & tag @WSUPullman #GoCougs https://t.co/EltXDy1tPtExample: Escaping double quotes using Twitter data
Using Twitter data you may encounter a lot of strings with double quotes.
- In the example below, our string includes special characters
\"and\nto escape the double quotes and the newline character.
#Twitter example of \" double quotes special characters p12_df$text[24] #> [1] "\"I really am glad that inside Engineering Student Services, I’ve been able to connect with my ESS advisor and professional development advisors there.\"\n-Alexandro Garcia, Civil & Environmental Engineering, 3rd year\n#imaberkeleyengineer #iamberkeley #voicesofberkeleyengineering https://t.co/ToVEynIUWH"- In the example below, our string includes special characters
Using
writeLines()we can see the contents of the strings as they would be read, rather than as R stores them.- We no longer see the escaped characters
\"or\n
writeLines(p12_df$text[24]) #> "I really am glad that inside Engineering Student Services, I’ve been able to connect with my ESS advisor and professional development advisors there." #> -Alexandro Garcia, Civil & Environmental Engineering, 3rd year #> #imaberkeleyengineer #iamberkeley #voicesofberkeleyengineering https://t.co/ToVEynIUWH- We no longer see the escaped characters