rm(list = ls()) # remove all objects in current environment
getwd()
#> [1] "/Users/jaquette/Documents/rclass1/lectures/patterns_base_r"
#load dataset with one obs per recruiting event
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_event_somevars.RData"))
#load("../../data/recruiting/recruit_event_somevars.Rdata")
#load dataset with one obs per high school
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_school_somevars.RData"))
#load("../../data/recruiting/recruit_school_somevars.Rdata")Investigating objects using Base R & subsetting with subset operators
1 Investigate objects, base R
Load .Rdata data frames we will use today
Data on off-campus recruiting events by public universities
- Data frame object
df_event- One observation per university, recruiting event
- Data frame object
df_school- One observation per high school (visited and non-visited)
1.1 Functions to describe objects
This section introduces some base R functions to describe objects (some of these you have seen before)
- list objects,
list.files()andls() - remove objects,
rm() - object type,
typeof() - object length (number of elements),
length() - object structure,
str() - number of rows and columns,
ncol()andnrow()
I use the functions typeof(), length(), str() anytime I encounter a new object
- Helps me understand the object before I start working with it
1.1.1 Listing objects
Files in your working directory
list.files() function lists files in your current working directory
- if you run this code from .qmd file, working directory is the location where the .qmd file is stored.
getwd() # what is your current working directory
#> [1] "/Users/jaquette/Documents/rclass1/lectures/patterns_base_r"
list.files()
#> [1] "base_r_week1_video_lecture_script.R" "fp1.JPG"
#> [3] "fp2.JPG" "one_carriage_train_vs_contents.png"
#> [5] "patterns_base_r_files" "patterns_base_r.html"
#> [7] "patterns_base_r.qmd" "patterns_base_r.rmarkdown"
#> [9] "smaller_trains.png" "test.txt"
#> [11] "three_carriage_train.png" "transform-logical.png"1.1.2 Objects currently open in your R session
Listing objects currently open in your R session
ls() function lists objects currently open in R
x <- "hello!"
ls() # Objects open in R
#> [1] "df_event" "df_school" "x"Removing objects currently open in your R session
rm() function removes specified objects open in R
rm(x)
ls()
#> [1] "df_event" "df_school"Command to remove all objects open in R (I don’t run it)
#rm(list = ls())1.1.3 Base R functions to describe objects, typeof()
typeof() function determines the the internal storage type of an object (e.g., logical vector, integer vector, list)
- syntax
tyepof(x)
- arguments
x: any R object
- help:
?typeofExamples
- Recall that a data frame is an object where type is a list
typeof(c(TRUE,TRUE,FALSE,NA))
#> [1] "logical"
typeof(df_event)
#> [1] "list"
typeof(x = df_event)
#> [1] "list"1.1.4 Base R functions to describe objects, length()
length() function determines the length of an R object
- for atomic vectors and lists,
length()is the number of elements in the object - syntax
length(x)
- arguments
x: any R object
- help:
?lengthExample, length of an atomic vector is
length(c(TRUE,TRUE,FALSE,NA))
#> [1] 4Example, length of a list or data frame
- length of a list is the number of elements
- data frame is a list
- length of a data frame = number of elements = number of variables
length(df_event) # = num elements = num columns
#> [1] 331.1.5 Base R functions to describe objects, str()
str() function compactly displays the structure of an R object
- “structure” includes type, length, and attribute of object and also nested objects
- syntax:
str(object) - arguments (partial)
object: any R objectmax.level: max level of nesting to display nested structures; defaultNA= all levels
- help:
?str
Example, atomic vectors
str(c(TRUE,TRUE,FALSE,NA))
#> logi [1:4] TRUE TRUE FALSE NA
str(object = c(TRUE,TRUE,FALSE,NA))
#> logi [1:4] TRUE TRUE FALSE NAExample, lists/data frames (output omitted)
x <- list(c(1,2), list("apple", "orange"), list(2, 3)) # list
str(x)
str(df_event) # data frame1.1.6 Base R functions to describe objects, ncol() and nrow()
ncol() nrow() and dim() functions
- Description
ncol()= number of columns;nrow()= number of rows
- syntax:
ncol(x)nrow(x)dim(x) - arguments
x: a vector, array, data frame, or NULL
- value/return:
- if object
xis an atomic vector:ncol()andnrow()returnsNULL - if object
xis a list but not a data frame:ncol()andnrow()returnsNULL - if object
xis a data frame:ncol()andnrow()returns integer of length 1
- if object
Example, object is a data frame
ncol(df_event) # num columns = num elements = num variables
#> [1] 33
nrow(df_event) # num rows = num observations
#> [1] 18680
# can wrap ncol() or nrow() within str() to see what functions return
#str(ncol(df_event))Example, object is atomic vector or list that is not a data frame (output omitted)
ncol(c(TRUE,TRUE,FALSE,NA)) # atomic vector
x <- list(c(1,2), list("apple", "orange"), list(2, 3)) # list
nrow(x)1.1.7 Base R functions to describe objects, dim()
dim() function returns the dimensions of an object (e.g., number of rows and columns)
- syntax:
dim(x) - arguments
x: a vector, array, data frame, or NULL
- value/return:
- if object
xis a data frame:dim()returns integer of length 2- first element = number of rows; second element = number of columns
- if object
xis an atomic vector:dim()returnsNULL - if object
xis a list but not a data frame:dim()returnsNULL
- if object
Example, object is a data frame
dim(df_event) # shows number rows by columns
#> [1] 18680 33
str(dim(df_event)) # can wrap dim() within str() to see what functions return
#> int [1:2] 18680 33Example, object is atomic vector or list that is not a data frame (output omitted)
dim(c(TRUE,TRUE,FALSE,NA)) # atomic vector
x <- list(c(1,2), list("apple", "orange"), list(2, 3)) # list
dim(x)1.2 Variables names
1.2.1 names() function
names() function gets or sets the names of elements of an object
- syntax:
- get the names of an object:
names(x) - set the names of an object:
names(x) <- value
- get the names of an object:
- arguments (partial)
x: an R objectvalue: a character vector with same length as objectxorNULL
- value/return
names(x)returns a character vector of length =length(x)in which each element is the name of the element ofx
Example, get names (of atomic vector)
a <- c(v1 = 1,v2 = 2, 3 ,v4 = "hi!") # named atomic vector
a
#> v1 v2 v4
#> "1" "2" "3" "hi!"
length(a)
#> [1] 4
names(a)
#> [1] "v1" "v2" "" "v4"
length(names(a)) # investigate length of object names(a)
#> [1] 4
str(names(a)) # investigate structure of object names(a)
#> chr [1:4] "v1" "v2" "" "v4"Example, set names (of atomic vector)
names(a) <- NULL # set names of vector a to NULL
a
#> [1] "1" "2" "3" "hi!"
names(a)
#> NULL
names(a) <- c("var1","var2","var3","var4") # set names of vector a
a
#> var1 var2 var3 var4
#> "1" "2" "3" "hi!"
names(a)
#> [1] "var1" "var2" "var3" "var4"1.2.2 Applying names() function to a data frame
Recall that a data frame is an object where type is a list and each element is named
- each element is a variable
- each element name is a variable name
Example (output omitted)
names(df_event)Investigate the object names(df_event)
typeof(names(df_event)) # type = character vector
#> [1] "character"
length(names(df_event)) # length = number of variables in data frame
#> [1] 33
str(names(df_event)) # structure of names(df_event)
#> chr [1:33] "instnm" "univ_id" "instst" "pid" "event_date" "event_type" ...We can even assign a new object based on names(df_event)
names_event <- names(df_event)
typeof(names_event) # type = character vector
#> [1] "character"
length(names_event) # length = number of variables in data frame
#> [1] 33
str(names_event) # structure of names(df_event)
#> chr [1:33] "instnm" "univ_id" "instst" "pid" "event_date" "event_type" ...1.2.3 Variable names
Refer to specific named elements of an object using this syntax:
object_name$element_name
When object is data frame, refer to specific variables using this syntax:
data_frame_name$varname- This approach to isolating variables is very useful for investigating data
#df_event$instnm
typeof(df_event$instnm)
#> [1] "character"
typeof(df_event$med_inc)
#> [1] "double"Data frames are lists with the following criteria:
- each element of the list is (usually) a vector; each element of a list is a variable
- length of data frame = number of variables
length(df_event)
#> [1] 33
nrow(df_event)
#> [1] 18680
#str(df_event)- each element of the list (i.e., variable) has the same length
- Length of each variable is equal to number of observations in data frame
typeof(df_event$event_state)
#> [1] "character"
length(df_event$event_state)
#> [1] 18680
str(df_event$event_state)
#> chr [1:18680] "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" ...
typeof(df_event$med_inc)
#> [1] "double"
length(df_event$med_inc)
#> [1] 18680
str(df_event$med_inc)
#> num [1:18680] 71714 89122 70136 70136 71024 ...The object df_school has one obs per high school
- variable
visits_by_100751shows number the of visits by University of Alabama to each high school - like all variables in a data frame, the var
visits_by_100751is just a vector
typeof(df_school$visits_by_100751)
#> [1] "integer"
length(df_school$visits_by_100751) # num elements in vector = num obs
#> [1] 21301
str(df_school$visits_by_100751)
#> int [1:21301] 0 0 0 0 0 0 0 0 0 0 ...
sum(df_school$visits_by_100751) # sum of values of var across all obs
#> [1] 3338We perform calculations on a variable like we would on any vector of same type
v <- c(2,4,6)
typeof(v)
#> [1] "double"
length(v)
#> [1] 3
sum(v)
#> [1] 121.3 View and print data
1.3.1 Viewing and printing, data frames
Many ways to view/print a data frame object. Here are three ways:
Simply type the object name (output omitted)
- number of observations and rows printed depend on YAML header settings and on object “attributes” (attributes discussed in future week)
df_event- Use the
View()function to view data in a browser
View(df_event)head()to show the first n rows. The default is 6 rows.
#?head
#head(df_event)
head(df_event, n=5)1.3.2 Viewing and printing, data frames
obj_name[<rows>,<cols>] to print specific rows and columns of data frame
- particularly powerful when combined with sequences (e.g.,
1:10)
Examples (output omitted):
- Print first five rows, all vars
df_event[1:5, ]- Print first five rows and first three columns
df_event[1:5, 1:3]- Print first three columns of the 100th observation
df_event[100, 1:3]- Print the 50th observation, all variables
df_event[50,]1.3.3 Viewing and printing, variables within data frames
Recall that:
obj_name$var_nameprint specifics elements (i.e., variables) of a data frame
df_event$zip- each element (i.e., variable) of data frame is an atomic vector with length = number of observations
typeof(df_event$zip)
#> [1] "character"
length(df_event$zip)
#> [1] 18680- each element of a variable is the value of the variable for one observation
Print specific elements (i.e., observations) of variable based on element position
- syntax:
obj_name$var_name[<element position>] - vectors don’t have “rows” or “columns”; they just have elements
- syntax combined with sequences (e.g., print first 10 observations)
df_event$event_state[1:10] # print obs 1-10 of variable "event_state"
#> [1] "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA" "MA"
df_event$event_type[6:10] # print obs 6-10 of variable "event_type"
#> [1] "private hs" "private hs" "public hs" "private hs" "public hs"Print specific elements (i.e., observations) of variable based on element position
- syntax:
obj_name$var_name[<element position>]
Example, print individual elements
df_event$zip[1:5] # print obs 1-5 of variable for event zip code
#> [1] "01002" "01007" "01020" "01020" "01027"
df_event$zip[1] # print obs 1 of variable for event zip code
#> [1] "01002"
df_event$zip[5] # print obs 5 of variable for event zip code
#> [1] "01027"
df_event$zip[c(1,3,5)] # print obs 1, 3, and 5 of variable for event zip code
#> [1] "01002" "01020" "01027"Print specific elements of multiple variables using combine function c()
- syntax:
c(obj_name$var1_name[<element position>], obj_name$var2_name[<element position>],...) - Example: print first five observations of variables
"event_state"and"event_type"
c(df_event$event_state[1:5],df_event$event_type[1:5])
#> [1] "MA" "MA" "MA" "MA" "MA" "public hs"
#> [7] "public hs" "public hs" "public hs" "public hs"1.3.4 Exercise
Printing exercise using the df_school data frame
- Use the
obj_name[<rows>,<cols>]syntax to print the first 5 rows and 3 columns of thedf_schooldata frame - Use the
head()function to print the first 4 observations - Use the
obj_name$var_name[1:10]syntax to print the first 10 observations of a variable in thedf_schooldata frame - Use combine() to print the first 3 observations of variables “school_type” & “name”
Solutions
- Use the
obj_name[<rows>,<cols>]syntax to print the first 5 rows and 3 columns of thedf_schooldata frame
df_school[1:5,1:3]
#> state_code school_type ncessch
#> 1 AK public 020000100208
#> 2 AK public 020000100211
#> 3 AK public 020000100212
#> 4 AK public 020000100213
#> 5 AK public 020000300216- Use the
head()function to print the first 4 observations
head(df_school, n=4)
#> state_code school_type ncessch name
#> 1 AK public 020000100208 Bethel Regional High School
#> 2 AK public 020000100211 Ayagina'ar Elitnaurvik
#> 3 AK public 020000100212 Kwigillingok School
#> 4 AK public 020000100213 Nelson Island Area School
#> address city zip_code pct_white pct_black
#> 1 1006 Ron Edwards Memorial Dr Bethel 99559 11.7764 0.5988
#> 2 106 Village Road Kongiganak 99559 0.0000 0.0000
#> 3 108 Village Road Kwigillingok 99622 0.0000 0.0000
#> 4 118 Village Road Toksook Bay 99637 0.0000 0.0000
#> pct_hispanic pct_asian pct_amerindian pct_other num_fr_lunch total_students
#> 1 1.5968 0.998 84.6307 0.3992 362 501
#> 2 0.0000 0.000 99.4505 0.5495 182 182
#> 3 0.0000 0.000 100.0000 0.0000 116 120
#> 4 0.0000 0.000 100.0000 0.0000 187 201
#> num_took_math num_prof_math num_took_rla num_prof_rla avgmedian_inc_2564
#> 1 146 24.82 147 24.99 76160.0
#> 2 17 1.70 17 1.70 76160.0
#> 3 14 3.50 14 3.50 NA
#> 4 30 3.00 30 3.00 57656.5
#> visits_by_110635 visits_by_126614 visits_by_100751 inst_110635 inst_126614
#> 1 0 0 0 CA CO
#> 2 0 0 0 CA CO
#> 3 0 0 0 CA CO
#> 4 0 0 0 CA CO
#> inst_100751
#> 1 AL
#> 2 AL
#> 3 AL
#> 4 AL- Use the
obj_name$var_name[1:10]syntax to print the first 10 observations of a variable in thedf_schooldata frame
df_school$name[1:10]
#> [1] "Bethel Regional High School" "Ayagina'ar Elitnaurvik"
#> [3] "Kwigillingok School" "Nelson Island Area School"
#> [5] "Alakanuk School" "Emmonak School"
#> [7] "Hooper Bay School" "Ignatius Beans School"
#> [9] "Pilot Station School" "Kotlik School"- Use combine() to print the first 3 observations of variables “school_type” & “name”
c(df_school$school_type[1:3],df_school$name[1:3])
#> [1] "public" "public"
#> [3] "public" "Bethel Regional High School"
#> [5] "Ayagina'ar Elitnaurvik" "Kwigillingok School"1.4 Missing values
1.4.1 Missing values
Missing values have the value NA
NAis a special keyword, not the same as the character string"NA"
use is.na() function to determine if a value is missing
is.na()returns a logical vector
is.na(5)
#> [1] FALSE
is.na(NA)
#> [1] TRUE
is.na("NA")
#> [1] FALSE
typeof(is.na("NA")) # example of a logical vector
#> [1] "logical"
nvector <- c(10,5,NA)
is.na(nvector)
#> [1] FALSE FALSE TRUE
typeof(is.na(nvector)) # example of a logical vector
#> [1] "logical"
svector <- c("e","f",NA,"NA")
is.na(svector)
#> [1] FALSE FALSE TRUE FALSE1.4.2 Missing values are “contagious”
What does “contagious” mean?
- operations involving a missing value will yield a missing value
7>5
#> [1] TRUE
7>NA
#> [1] NA
sum(1,2,NA)
#> [1] NA
0==NA
#> [1] NA
2*c(0,1,2,NA)
#> [1] 0 2 4 NA
NA*c(0,1,2,NA)
#> [1] NA NA NA NA1.4.3 Functions and missing values example, table()
table() function is useful for investigating categorical variables
str(df_event$event_type)
#> chr [1:18680] "public hs" "public hs" "public hs" "public hs" "public hs" ...
table(df_event$event_type)
#>
#> 2yr college 4yr college other private hs public hs
#> 951 531 2001 3774 11423By default table() ignores NA values
#?table
str(df_event$school_type_pri)
#> int [1:18680] NA NA NA NA NA 1 1 NA 1 NA ...
table(df_event$school_type_pri)
#>
#> 1 2 5
#> 3765 8 1useNA argument controls if table includes counts of NAs. Allowed values:
- never (“no”) [DEFAULT VALUE]
- only if count is positive (“ifany”);
- even for zero counts (“always”)”
nrow(df_event)
#> [1] 18680
table(df_event$school_type_pri, useNA="always")
#>
#> 1 2 5 <NA>
#> 3765 8 1 14906Broader point: Most functions that create descriptive statistics have options about how to treat missing values`
- When investigating data, good practice to always show missing values
2 Subsetting using subset operators
2.0.1 Subsetting to Extract Elements
“Subsetting” refers to isolating particular elements of an object
Subsetting operators can be used to select/exclude elements (e.g., variables, observations)
- there are three subsetting operators:
[],$,[[]] - these operators function differently based on vector types (e.g, atomic vectors, lists, data frames)
2.0.2 Wichham refers to number of “dimensions” in R objects
An atomic vector is a 1-dimensional object that contains n elements
x <- c(1.1, 2.2, 3.3, 4.4, 5.5)
str(x)
#> num [1:5] 1.1 2.2 3.3 4.4 5.5Lists are multi-dimensional objects
- Contains n elements; each element may contain a 1-dimensional atomic vector or a multi-dimensional list. Below list contains 3 dimensions
list <- list(c(1,2), list("apple", "orange"))
str(list)
#> List of 2
#> $ : num [1:2] 1 2
#> $ :List of 2
#> ..$ : chr "apple"
#> ..$ : chr "orange"Data frames are 2-dimensional lists
- each element is a variable (dimension=columns)
- within each variable, each element is an observation (dimension=rows)
ncol(df_school)
#> [1] 26
nrow(df_school)
#> [1] 213012.1 Subset atomic vectors using []
“Subsetting” a vector refers to isolating particular elements of a vector
- I sometimes refer to this as “accessing elements of a vector”
- subsetting elements of a vector is similar to “filtering” rows of a data-frame
[]is the subsetting function for vectors
Six ways to subset an atomic vector using []
- Using positive integers to return elements at specified positions
- Using negative integers to exclude elements at specified positions
- Using logicals to return elements where corresponding logical is
TRUE - Empty
[]returns original vector (useful for dataframes) - Zero vector [0], useful for testing data
- If vector is “named,” use character vectors to return elements with matching names
2.1.1 1. Using positive integers to return elements at specified positions (subset atomic vectors using [])
Create atomic vector x
(x <- c(1.1, 2.2, 3.3, 4.4, 5.5))
#> [1] 1.1 2.2 3.3 4.4 5.5
str(x)
#> num [1:5] 1.1 2.2 3.3 4.4 5.5[] is the subsetting function for vectors
- contents inside
[]can refer to element number (also called “position”).- e.g.,
[3]refers to contents of 3rd element (or position 3)
- e.g.,
x[5] #return 5th element
#> [1] 5.5
x[c(3, 1)] #return 3rd and 1st element
#> [1] 3.3 1.1
x[c(4,4,4)] #return 4th element, 4th element, and 4th element
#> [1] 4.4 4.4 4.4
#Return 3rd through 5th element
x[3:5]
#> [1] 3.3 4.4 5.52.1.2 2. Using negative integers to exclude elements at specified positions (subset atomic vectors using [])
Before excluding elements based on position, investigate object
x
#> [1] 1.1 2.2 3.3 4.4 5.5
length(x)
#> [1] 5
str(x)
#> num [1:5] 1.1 2.2 3.3 4.4 5.5Use negative integers to exclude elements based on element position
x[-1] # exclude 1st element
#> [1] 2.2 3.3 4.4 5.5
x[c(3,1)] # 3rd and 1st element
#> [1] 3.3 1.1
x[-c(3,1)] # exclude 3rd and 1st element
#> [1] 2.2 4.4 5.52.1.3 3. Using logicals to return elements where corresponding logical is TRUE (subset atomic vectors using [])
x
#> [1] 1.1 2.2 3.3 4.4 5.5When using x[y] to subset x, good practice to have length(x)==length(y)
length(x) # length of vector x
#> [1] 5
length(c(TRUE,FALSE,TRUE,FALSE,TRUE)) # length of y
#> [1] 5
length(x) == length(c(TRUE,FALSE,TRUE,FALSE,TRUE)) # condition true
#> [1] TRUE
x[c(TRUE,TRUE,FALSE,FALSE,TRUE)]
#> [1] 1.1 2.2 5.5Recycling rules:
- in
x[y], ifxis different length thany, R “recycles” length of shorter to match length of longer
length(c(TRUE,FALSE))
#> [1] 2
x
#> [1] 1.1 2.2 3.3 4.4 5.5
x[c(TRUE,FALSE)]
#> [1] 1.1 3.3 5.5x
#> [1] 1.1 2.2 3.3 4.4 5.5Note that a missing value (NA) in the index always yields a missing value in the output:
x[c(TRUE, FALSE, NA, TRUE, NA)]
#> [1] 1.1 NA 4.4 NAReturn all elements of object x where element is greater than 3:
x # print object X
#> [1] 1.1 2.2 3.3 4.4 5.5
x>3 # for each element of X, print T/F whether element value > 3
#> [1] FALSE FALSE TRUE TRUE TRUE
str(x>3)
#> logi [1:5] FALSE FALSE TRUE TRUE TRUE
x[x>3] # prints only the values that had TRUE at that position
#> [1] 3.3 4.4 5.5The visits_by_100751 column shows how many visits the University of Alabama made to each school. Let’s subset this to only include 2 or more visits:
df_school$visits_by_100751[1:100]
#> [1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
#> [38] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
#> [75] 0 0 0 0 0 0 0 0 0 0 5 2 4 4 3 3 3 3 3 3 2 3 3 3 3 1
df_school$visits_by_100751[1:100]>2
#> [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [49] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [61] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [73] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [85] TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE
#> [97] TRUE TRUE TRUE FALSE
df_school$visits_by_100751[df_school$visits_by_100751>2]
#> [1] 5 4 4 3 3 3 3 3 3 3 3 3 3 4 4 3 3 3 3 3 4 4 3 3 3 4 3 3 3 3 3 3 3 4 3 3 4
#> [38] 4 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 4 3 3 3 3 4 5 3 3 3 4 3 3 4 3 3 3 3
#> [75] 3 4 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 5 4 6 5 3 5 5 5 3
#> [112] 4 4 3 3 3 5 3 4 4 3 3 3 3 4 4 3 3 4 3 3 3 3 3 3 3 3 3 3 3 4 3 3 3 3 3 3 3
#> [149] 3 3 3 3 4 3 3 3 3 3 3 3 32.1.4 4. Empty [] returns original vector (subset atomic vectors using [])
x
#> [1] 1.1 2.2 3.3 4.4 5.5
x[]
#> [1] 1.1 2.2 3.3 4.4 5.5This is useful for sub-setting data frames, as we will show below
2.1.5 5. Zero vector [0] (subset atomic vectors using [])
Zero vector, x[0]
- R interprets this as returning element 0
x[0]
#> numeric(0)Wickham states:
- “This is not something you usually do on purpose, but it can be helpful for generating test data.”
2.1.6 6. If vector is named, character vectors to return elements with matching names (subset atomic vectors using [])
Create vector y that has values of vector x but each element is named
x
#> [1] 1.1 2.2 3.3 4.4 5.5
(y <- c(a=1.1, b=2.2, c=3.3, d=4.4, e=5.5))
#> a b c d e
#> 1.1 2.2 3.3 4.4 5.5Return elements of vector based on name of element
- enclose element names in single
''or double""quotes
#show element named "a"
y["a"]
#> a
#> 1.1
#show elements "a", "b", and "d"
y[c("a", "b", "d" )]
#> a b d
#> 1.1 2.2 4.42.2 Subsetting lists/data frames using []
Using [] operator to subset lists works the same as subsetting atomic vector
- Using
[]with a list always returns a list
list_a <- list(list(1,2),3,"apple")
str(list_a)
#> List of 3
#> $ :List of 2
#> ..$ : num 1
#> ..$ : num 2
#> $ : num 3
#> $ : chr "apple"
#create new list that consists of elements 3 and 1 of list_a
list_b <- list_a[c(3, 1)]
str(list_b)
#> List of 2
#> $ : chr "apple"
#> $ :List of 2
#> ..$ : num 1
#> ..$ : num 2
#show elements 3 and 1 of object list_a
#str(list_a[c(3, 1)])Recall that a data frame is just a particular kind of list
- each element = a column = a variable
Using [] with a list always returns a list
- Using
[]with a data frame always returns a data frame
Two ways to use [] to extract elements of a data frame
- use “single index”
df_name[<columns>]to extract columns (variables) based on element position number (i.e., column number) - use “double index”
df_name[<rows>, <columns>]to extact particular rows and columns of a data frame
2.2.1 Subsetting data frames using [] to extract columns (variables) based on element position
Use “single index” df_name[<columns>] to extract columns (variables) based on element number (i.e., column number)
Examples [output omitted]
names(df_event)
#extract elements 1 through 4 (elements=columns=variables)
df_event[1:4]
df_event[c(1,2,3,4)]
str(df_event[1:4])
#extract columns 13 and 7
df_event[c(13,7)]2.2.2 Subsetting Data Frames to extract columns (variables) and rows (observations) based on positionality
use “double index” syntax df_name[<rows>, <columns>] to extact particular rows and columns of a data frame
- often combined with sequences (e.g.,
1:10)
#Return rows 1-3 and columns 1-4
df_event[1:3, 1:4]
#> instnm univ_id instst pid
#> 1 UM Amherst 166629 MA 57570
#> 2 UM Amherst 166629 MA 56984
#> 3 UM Amherst 166629 MA 57105
#Return rows 50-52 and columns 10 and 20
df_event[50:52, c(10,20)]
#> event_state pct_tworaces_zip
#> 50 MA 1.9831
#> 51 MA 1.9831
#> 52 MA 1.9831Use “double index” syntax df_name[<rows>, <columns>] to extact particular rows and columns of a data frame
recall that empty [] returns original object (output omitted)
#return original data frame
df_event[]
#return specific rows and all columns (variables)
df_event[1:5, ]
#return all rows and specific columns (variables)
df_event[, c(1,2,3)]2.2.3 Use [] to extract data frame columns based on variable names
Selecting columns from a data frame by subsetting with [] and list of element names (i.e., variable names) enclose in quotes
“single index” approach extracts specific variables, all rows (output omitted)
df_event[c("instnm", "univ_id", "event_state")] “Double index” approach extracts specific variables and specific rows
- syntax
df_name[<rows>, <columns>]
df_event[1:5, c("instnm", "event_state", "event_type")]
#> instnm event_state event_type
#> 1 UM Amherst MA public hs
#> 2 UM Amherst MA public hs
#> 3 UM Amherst MA public hs
#> 4 UM Amherst MA public hs
#> 5 Stony Brook MA public hs2.2.4 Student exercises
Use subsetting operators from base R in extracting columns (variables), observations:
Use both “single index” and “double index” in subsetting to create a new dataframe by extracting the columns
instnm,event_date,event_typefrom thedf_eventdata frame. And show what columns (variables) are in the newly created dataframe.Use subsetting to return rows 1-5 of columns
state_code,name,addressfrom thedf_schooldata frame.
Solutions
Solution to 1
base R using subsetting operators
# single index
df_event_br <- df_event[c("instnm", "event_date", "event_type")]
#double index
df_event_br <- df_event[, c("instnm", "event_date", "event_type")]
names(df_event_br)
#> [1] "instnm" "event_date" "event_type"Solution to 2
base R using subsetting operators
df_school[1:5, c("state_code", "name", "address")]
#> state_code name address
#> 1 AK Bethel Regional High School 1006 Ron Edwards Memorial Dr
#> 2 AK Ayagina'ar Elitnaurvik 106 Village Road
#> 3 AK Kwigillingok School 108 Village Road
#> 4 AK Nelson Island Area School 118 Village Road
#> 5 AK Alakanuk School 9 School Road2.3 Subsetting lists/data frames using [[]] and $
2.3.1 Subset single element from object using [[]] operator, atomic vectors
So far we have used [] to extract elements from an object
- Apply
[]to atomic vector: returns atomic vector with elements you requested - Apply
[]to list: returns list with elements you requested
[[]] also extract elements from an object
- Applying
[[]]to atomic vector gives same result as[]; that is, an atomic vector with element you request
(x <- c(1.1, 2.2, 3.3, 4.4, 5.5))
#> [1] 1.1 2.2 3.3 4.4 5.5
str(x[3])
#> num 3.3
str(x[[3]])
#> num 3.3- Caveat: when applying
[[]]to atomic vector, you can only subset a single element
x[c(3,4)] # single bracket; this works
#> [1] 3.3 4.4
#x[[c(3,4)]] # double bracket; this won't work2.3.2 Subsetting lists using [] vs. [[]], introduce “train metaphor”
Applying [[]] to a list
- Understanding what
[]vs.[[]]does to a list is very important but requires some explanation!
Advanced R chapter 4.3 by Wickham uses the “train metaphor” to explain a list vs. contents of a list and how this relates to [] vs. [[]]
Below code chunk makes a list named list_x that contains 3 elements
list_x <- list(1:3, "a", 4:6) # create list object list_x
str(list_x)
#> List of 3
#> $ : int [1:3] 1 2 3
#> $ : chr "a"
#> $ : int [1:3] 4 5 6In our train metaphor, object list_x is a train that contains 3 carriages
list object list_x is a train that contains 3 carriages
#> Warning: package 'knitr' was built under R version 4.3.3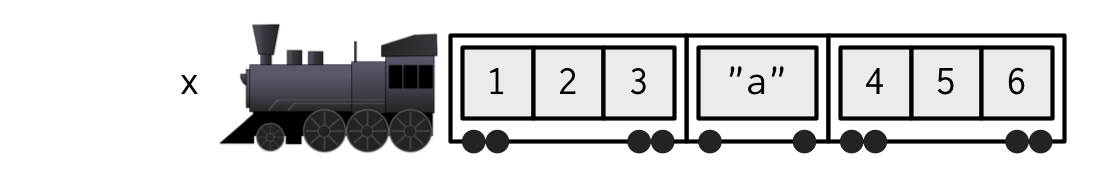
When we “subset a list” – that is, extract one or more elements from the list – we have two broad choices (image below)
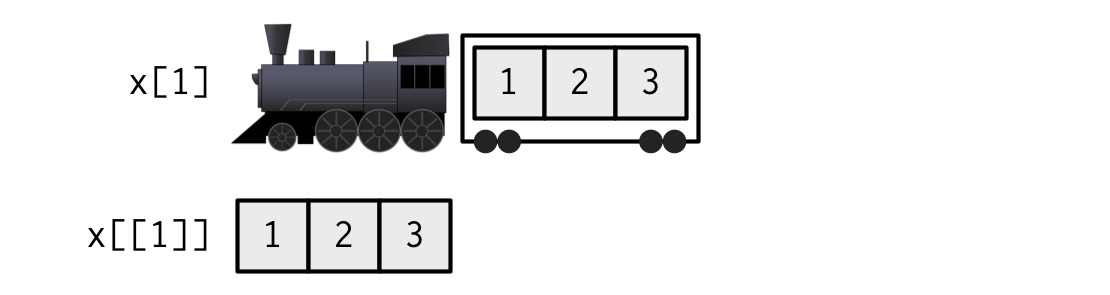
Extracting elements using
[]always returns a list, usually one with fewer elements- you can think of this as a train with fewer carriages
#str(list_x) str(list_x[1]) # returns a list #> List of 1 #> $ : int [1:3] 1 2 3Extracting element using
[[]]returns contents of particular carriage- I say applying
[[]]to a list or data frame returns a simpler object that moves up one level of hierarchy
str(list_x[[1]]) # returns an atomic vector #> int [1:3] 1 2 3- I say applying
2.3.3 Subset lists using [] vs. [[]], deepen understanding of []
Rules about applying subset operator [] to a list
- Applying
[]to a list always returns a list - Resulting list contains 1 or more elements depending on what typed inside
[]
Here is a list object named list_x
list_x <- list(1:3, "a", 4:6)Here is an image of a few “trains” that can be created by applying [] to list_x
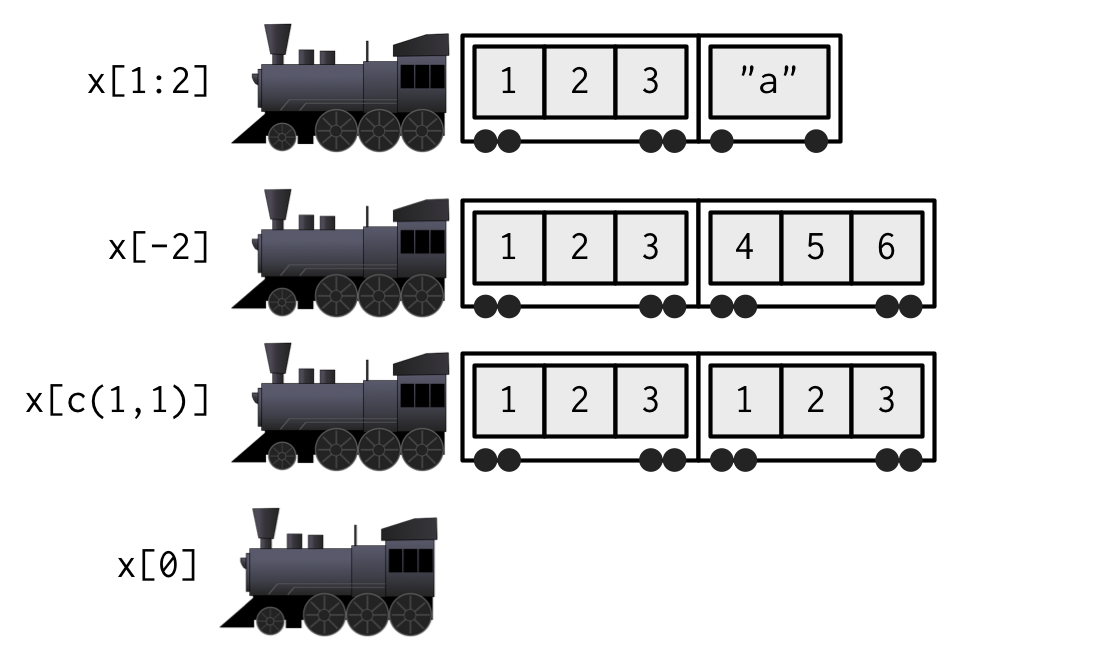
And here is code to create the “trains” shown in above image (output omitted)
list_x[1:2]
list_x[-2]
list_x[c(1,1)]
list_x[0]
list_x[] # returns the original list; not shown in above train pictureRules about applying subset operator [[]] to a list
- Can apply
[[]]to return the contents of a single element of a list
Create list list_x and show “train” Image of applying list_x[1] vs. list_x[[1]]
list_x <- list(1:3, "a", 4:6)
Object created by list_x[1] is a list with one element (output omitted)
list_x[1]
#> [[1]]
#> [1] 1 2 3
str(list_x[1])
#> List of 1
#> $ : int [1:3] 1 2 3Object created by list_x[[1]] is a vector with 3 elements (output omitted)
list_x[[1]]gives us “contents” of element 1- Since element 1 contains a numeric vector, object created by
list_x[[1]]is a numeric vector
list_x[[1]]
#> [1] 1 2 3
str(list_x[[1]])
#> int [1:3] 1 2 3Rules about applying subset operator [[]] to a list
- Can apply
[[]]to return the contents of a single element of a list
list_x <- list(1:3, "a", 4:6) # create list list_xWe cannot use [[]] to subset multiple elements of a list (output omitted)
- e.g., we could write
list_x[[2]]but notlist_x[[2:3]]
list_x[[c(2)]] # this works, subset element 2 using [[]]
list_x[[c(2,3)]] # this doesn't work; subset element 2 and 3 using [[]]
list_x[c(2,3)] # this works; subset element 2 and 3 using []Like [], can use [[]] to return contents of named elements specified using quotes
- syntax:
obj_name[["element_name"]]
list_x <- list(var1=1:3, var2="a", var3=4:6) # create list with named elementsSubset list list_x using [[]] element names
list_x[["var1"]] # subset by element position: list_x[[1]]
#> [1] 1 2 3
str(list_x[["var1"]])
#> int [1:3] 1 2 3
str(list_x["var1"]) # note: suggests var name is attribute of list, not atomic vector
#> List of 1
#> $ var1: int [1:3] 1 2 3Can do same thing with data frames because data frames are lists (output omitted)
- e.g.,
df_event[["zip"]]returns contents of element named"zip" - object created by
df_event[["zip"]]is character vector of length = 18,680
# df_event[["zip"]] # this works but long output
str(df_event[["zip"]]) # character vector of length 18,860
typeof(df_event[["zip"]])
length(df_event[["zip"]])
str(df_event["zip"]) # by contrast, this is a dataframe w/ one variable2.3.4 General rules of applying [] vs [[]] to (nested) objects
What we just learned about applying [] vs [[]] to lists applies more generally to “nested objects”
- “nested objects” are objects with a hierarchical structure such that an element of an object contains another object
General rules of applying [] vs. [[]] to nested objects
- subset any object
xusing[]will return object with same data structure asx - subset any object
xusing[[]]will return an object thay may or may not have same data structure ofx- if object
xis not a nested object, then applying[[]]to a single element ofxwill return object with same data structure asx - if object
xhas a nested data structure, then then applying[[]]to a single element ofxwill “move up one level of hierarchy” to extract the contents of an element within the objectx
- if object
When working w/ data frames, functions that calculate things expect to be working with atomic vectors (think [[]]) not lists (think [])
mean(df_event[['med_inc']], na.rm = TRUE)
#> [1] 89089.28
# mean(df_event['med_inc'], na.rm = TRUE) # by contrast, this doesn't work
str(df_event['med_inc'])
#> Classes 'tbl_df', 'tbl' and 'data.frame': 18680 obs. of 1 variable:
#> $ med_inc: num 71714 89122 70136 70136 71024 ...
str(df_event[['med_inc']])
#> num [1:18680] 71714 89122 70136 70136 71024 ...2.3.5 Subset lists/data frames using $
list_x <- list(var1=1:3, var2="a", var3=4:6)obj_name$element_name is shorthand operator for obj_name[["element_name"]]
These three lines of code all give the same result
list_x[[1]]
#> [1] 1 2 3
list_x[["var1"]]
#> [1] 1 2 3
list_x$var1
#> [1] 1 2 3df_name$var_name: easiest way in base R to refer to variable in a data frame
- these two lines of code are equivalent
str(df_event[["zip"]])
#> chr [1:18680] "01002" "01007" "01020" "01020" "01027" "01027" "01027" ...
str(df_event$zip)
#> chr [1:18680] "01002" "01007" "01020" "01020" "01027" "01027" "01027" ...3 Appendix
3.1 Subset Data frames by combining [] and $
Motivation
- When working with data frames we often want to isolate those observations that satisfy certain conditions
- This is often referred to as “filtering”
- We filter observations based on the values of one or more variables
- Perhaps you have seen “filtering” in Microsoft Excel
- open some spreadsheet that contains variables (columns) and observations (rows)
- click on
Data>>Filterand then filter observations based on values of variable(s)
Filtering example using data frame df_school
- Observations:
- One observation per high school (public and private)
- Variables:
- high school characteristics; number of off-campus recruiting visits from particular universities
- NCES ID for UC Berkeley is
110635 - variable
visits_by_110635shows number of visits a high school received from UC Berkeley
Task:
- Isolate obs where school received at least 1 visit from UC Berkeley
General syntax: df_name[df_name$var_name <condition>, ]
- where
<condition>is something that evaluates toTRUEorFALSEfor each element of the atomic vector (i.e., variable) - Note that syntax uses “double index”
df_name[<rows>, <columns>]syntax- Therefore, the
<condition>in above syntax is isolating<rows>
- Therefore, the
- Cannot use “single index” syntax
df_name[<columns>]
Solution to task (output omitted)
- Note: below code filters observations but keeps all variables
df_school[df_school$visits_by_110635 >= 1, ]3.1.1 Subset Data Frames by combining [] and $, decompose syntax
Task: Isolate obs where school received at least 1 visit from UC Berkeley
- general syntax:
df_name[df_name$var_name <condition>, ] - solution:
df_school[df_school$visits_by_110635 >= 1, ]
Decomposing syntax df_school[df_school$visits_by_110635 >= 1, ]
df_school$visits_by_110635 >= 1: returns a logical (TRUE/FALSE) atomic vector with length equal to number of obs indf_school
typeof(df_school$visits_by_110635 >= 1)
length(df_school$visits_by_110635 >= 1)
str(df_school$visits_by_110635 >= 1)df_school[df_school$visits_by_110635 >= 1, ]- uses “double index”
df_name[<rows>, <columns>]syntax to extract rows, columns - rows: extract rows where
df_school$visits_by_110635 >= 1isTRUE - columns: since
<columns>is empty, extracts all columns
- uses “double index”
- key point:
df_name[df_name$var_name <condition>, ]is “subset a vector approach #3”: “Using logicals to return elements where conditionTRUE” - example using atomic vectors (output omitted)
x <- c(1.1, 2.2, 3.3, 4.4, 5.5)
x[x>3]3.1.2 Subset Data Frames by combining [] and $, keep desired columns
General syntax to filter desired observations (rows) and variables (columns) of data frame:
df_name[df_name$var_name <condition>, <desired columns>]
Tasks (output omitted)
- Extract observations where the high school received at least 1 visit from UC Berkeley and the first three columns
df_school[df_school$visits_by_110635 >= 1, 1:3]- Extract observations where the high school received at least 1 visit from UC Berkeley and variables “state_code” “school_type” “name”
df_school[df_school$visits_by_110635 >= 1, c("state_code","school_type","name")]3.1.3 Subset Data Frames by combining [] and $, more examples
Syntax:
- filter based on one variable:
df_name[df_name$var_name <condition>, <columns>]
- Example syntax to filter based on two conditions being true
df_name[df_name$var_name <condition> & df_name$var_name <condition>, <columns>]
Pro tip:
- wrap above syntax within
nrow()function to count how many observations (rows) satisfy the condition (as opposed to printing all rows that satisfy condition)
Tasks
- Count obs where high schools received at least 1 visit by Bama (100751) and at least one visit by Berkeley (110635)
nrow(df_school[df_school$visits_by_110635 >= 1 &
df_school$visits_by_100751 >= 1, ])
#> [1] 247
# Equivalently:
# nrow(df_school[df_school[["visits_by_110635"]] >= 1 &
# df_school[["visits_by_100751"]] >= 1, ])- Count obs where schools received 1+ visit by Bama or 1+ visit by Berkeley
nrow(df_school[df_school$visits_by_110635 >= 1
| df_school$visits_by_100751 >= 1, ])
#> [1] 27633.1.4 Logical operators for comparisons
- Logical operators to isolate/filter observations of data frame
| Symbol | Meaning |
|---|---|
== |
Equal to |
!= |
Not equal to |
> |
greater than |
>= |
greater than or equal to |
< |
less than |
<= |
less than or equal to |
& |
AND |
| |
OR |
%in% |
includes |
- Visualization of “Boolean” operators (e.g., AND, OR, AND NOT)
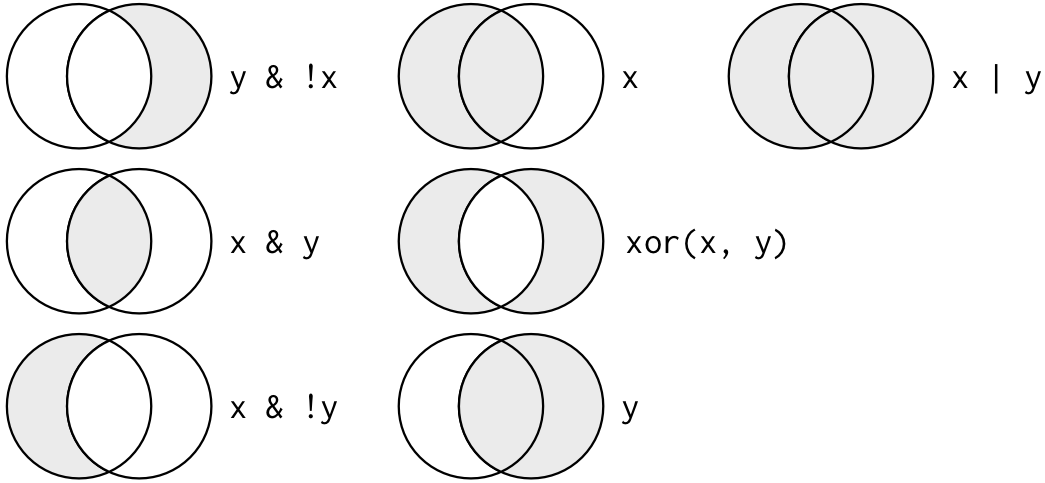
Example: Count the number of out-of-state high schools that UC Berkeley visited.
# The `inst_110635` variable contains the home state of UC Berkeley
unique(df_school$inst_110635)
#> [1] "CA"
# Filter for schools visited by UC Berkeley AND whose state is not "CA"
nrow(df_school[df_school$visits_by_110635 >= 1 &
df_school$state_code != df_school$inst_110635, ])
#> [1] 302Example: Count the number of schools in the Northeast that received a visit from either UC Berkeley, U of Alabama, or CU Boulder.
# Vector containing states located in the Northeast region
northeast_states <- c('CT', 'ME', 'MA', 'NH', 'RI', 'VT', 'NJ', 'NY', 'PA')
# Filter for schools in the Northeast AND visited by any of the 3 univs
nrow(df_school[df_school$state_code %in% northeast_states &
(df_school$visits_by_110635 >= 1 |
df_school$visits_by_100751 >= 1 |
df_school$visits_by_126614 >= 1), ])
#> [1] 5443.1.5 Subset Data Frames by combining [] and $, NA Observations
Filtering observations of data frame using [] combined with $ is more complicated in the presence of missing values (NA values)
The below we will explain
- why it is more complicated
- how to filter correctly when
NAs are present
When sub-setting via [] combined with $, result will include:
- rows where condition is
TRUE - as well as rows with
NA(missing) values for<condition>.
Task (using df_event, which has one obs per university, recruiting event)
- How many events at public HS with at least $50k median household income?
sum(is.na(df_event$med_inc)) # number of observations (all events) with missing values for med_inc
#> [1] 470
#num obs event_type=="public hs" and med_inc is missing
nrow(df_event[df_event$event_type == "public hs"
& is.na(df_event$med_inc)==1 , ]) # note TRUE evaluates to 1
#> [1] 75
#num obs event_type=="public hs" & med_inc is not NA & med_inc >= $50,000
nrow(df_event[df_event$event_type == "public hs"
& is.na(df_event$med_inc)==0 & df_event$med_inc>=50000 , ]) # note FALSE evaluates to 0
#> [1] 9941
#num obs event_type=="public hs" and med_inc >= $50,000
nrow(df_event[df_event$event_type == "public hs"
& df_event$med_inc>=50000 , ])
#> [1] 10016To exclude rows where condition is NA if subset using [] combined w/ $
- use
which()to ask only for values where condition evaluates toTRUE which()returns position numbers for elements where condition isTRUE
#?which
c(TRUE,FALSE,NA,TRUE)
#> [1] TRUE FALSE NA TRUE
str(c(TRUE,FALSE,NA,TRUE))
#> logi [1:4] TRUE FALSE NA TRUE
which(c(TRUE,FALSE,NA,TRUE))
#> [1] 1 4Task: Count events at public HS with at least $50k median household income?
#Base R, `[]` combined with `$`; without which()
nrow(df_event[df_event$event_type == "public hs" & df_event$med_inc>=50000, ])
#> [1] 10016
#Base R, `[]` combined with `$`; with which()
nrow(df_event[which(df_event$event_type == "public hs"
& df_event$med_inc>=50000), ])
#> [1] 99413.1.6 Student Exercises
Subsetting Data Frames with [] and $:
Show how many public high schools in California with at least 50% Latinx (hispanic in data) student enrollment from the dataframe df_school.
Show how many out-state events at public high schools with more than $30K median from the datframe df_event (do not forget to exclude missing values).
Solutions
Solution to 1
base R using [] and $
df_school_br1<- df_school[df_school$school_type == "public"
& df_school$pct_hispanic >= 50
& df_school$state_code == "CA", ]
nrow(df_school_br1)
#> [1] 713Solution to 2:
base R using [] and $
# use is.na to exclude NA
nrow(df_event[df_event$event_type == "public hs" & df_event$event_inst =="Out-State"
& df_event$med_inc > 30000 & is.na(df_event$med_inc) ==0, ])
#> [1] 7784
# use which to exclude NA
nrow(df_event[which(df_event$event_type == "public hs" & df_event$event_inst =="Out-State"
& df_event$med_inc > 30000 ), ])
#> [1] 77843.2 Subset using subset() function
The subset() is a base R function to “filter” observations from some object x
- object
xcan be a matrix, data frame, list subset()automatically excludes elements/rows withNAfor condition- Can also use
subset()to select variables - what
subset()function returns:- “An object similar to x contain just the selected rows and columns (for a matrix or data frame)”
subset()can be combined with:- assignment (
<-) to create new objects nrow()to count number of observations that satisfy criteria
- assignment (
?subsetSyntax [when object is data frame]: subset(x, subset, select, drop = FALSE)
xis object to be subsetsubsetis the logical expression(s) (evaluates toTRUE/FALSE) indicating elements (rows) to keepselectindicates columns to select from data frame (if argument is not used default will keep all columns)dropto preserve original dimensions [SKIP]
3.2.1 Subset function, examples
Recall the previous example where we count events at public HS with at least $50k median household income.
- Note.
subset()automatically excludes rows where condition isNA:
#Base R, `[]` combined with `$`, without which(); includes `NA`
nrow(df_event[df_event$event_type == "public hs"
& df_event$med_inc>=50000, ])
#> [1] 10016
#Base R, `[]` combined with `$`, with which(); excludes `NA`
nrow(df_event[which(df_event$event_type == "public hs"
& df_event$med_inc>=50000), ])
#> [1] 9941
#Base R, `subset()`; excludes `NA`
nrow(subset(df_event, event_type == "public hs"
& med_inc>=50000))
#> [1] 9941
#Base R, `subset()`; excludes `NA`; explicitly name arguments of subset()
nrow(subset(x = df_event, subset = event_type == "public hs"
& med_inc>=50000))
#> [1] 9941Using df_school, show all public high schools that are at least 50% Latinx (var=pct_hispanic) student enrollment in California
- Using base R,
subset()[output omitted]
#public high schools with at least 50% Latinx student enrollment
subset(x= df_school, subset = school_type == "public" & pct_hispanic >= 50
& state_code == "CA")- Can wrap
subset()withinnrow()to count number of observations that satisfy criteria
nrow(subset(df_school, school_type == "public" & pct_hispanic >= 50
& state_code == "CA"))
#> [1] 713Note that subset() identify the number of observations for which the condition is TRUE
nrow(subset(x = df_school, subset = TRUE))
#> [1] 21301
nrow(subset(x = df_school, subset = FALSE))
#> [1] 0Count all CA public high schools that are at least 50% Latinx and received at least 1 visit from UC Berkeley (var=visits_by_110635)
nrow(subset(df_school, school_type == "public" & pct_hispanic >= 50
& state_code == "CA" & visits_by_110635 >= 1))
#> [1] 100subset() can also use %in% operator, which is more efficient version of OR operator |
- Count number of schools from MA, ME, or VT that received at least one visit from University of Alabama (var=
visits_by_100751)
nrow(subset(df_school, state_code %in% c("MA","ME","VT")
& visits_by_100751 >= 1))
#> [1] 108Use the select argument within subset() to keep selected variables
- syntax:
select = c(var_name1,var_name2,...,var_name_n)
Subset all CA public high schools that are at least 50% Latinx AND only keep variables name and address
nrow(subset(x = df_school, subset = school_type == "public" & pct_hispanic >= 50
& state_code == "CA", select = c(name, address)))
#> [1] 713Combine subset() with assignment (<-) to create a new data frame
Create a new date frame of all CA public high schools that are at least 50% Latinx AND only keep variables name and address
df_school_v2 <- subset(df_school, school_type == "public" & pct_hispanic >= 50
& state_code == "CA", select = c(name, address))
head(df_school_v2, n=5)
#> name address
#> 1254 Tustin High 1171 El Camino Real
#> 1301 Bell Gardens High 6119 Agra St.
#> 1309 Santa Ana High 520 W. Walnut
#> 1332 Warren High 8141 De Palma St.
#> 1336 Hollywood Senior High 1521 N. Highland Ave.
nrow(df_school_v2)
#> [1] 7133.2.2 Student Exercises
Using subset() from base R:
Create a new dataframe by extracting the columns
instnm,event_date,event_typefromdf_eventdata frame. And show what columns (variables) are in the newly created dataframe.Create a new dataframe from the
df_schooldata frame that includes out-of-state public high schools with 50%+ Latinx student enrollment that received at least one visit by the University of California Berkeley (var= visits_by_110635). And count the number of observations.Count the number of public schools from CA, FL or MA that received one or two visits from UC Berkeley from the
df_schooldata frame.Subset all public out-of-state high schools visited by University of California Berkeley that enroll at least 50% Black students, and only keep variables
state_code,nameandzip_code.
Solutions
Solution to 1
df_event_br <- subset(df_event, select=c(instnm, event_date, event_type))
names(df_event_br)
#> [1] "instnm" "event_date" "event_type"Solution to 2
df_school_br <- subset(df_school, state_code != "CA" & school_type == "public"
& pct_hispanic >= 50 & visits_by_110635 >=1 )
nrow(df_school_br)
#> [1] 10Solution to 3
nrow(subset(df_school, state_code %in% c("CA", "FL", "MA")
& school_type == "public" & visits_by_110635 %in% c(1,2) ))
#> [1] 246Solution to 4
subset(df_school, school_type == "public" & state_code != "CA"
& visits_by_110635 >= 1 & pct_black >= 50,
select = c(state_code, name, zip_code))
#> state_code name zip_code
#> 4523 GA Grady High School 30309
#> 8519 MD Frederick Douglass High 20772
#> 9666 MN DOWNTOWN CAMPUS 55403
#> 10681 MS MURRAH HIGH SCHOOL 39202
#> 14569 OH Shaker Hts High School 44120
#> 14632 OH Cleveland Heights High School 44118
#> 17142 SC Spring Valley High 29229
#> 17143 SC Richland Northeast High 29223
#> 17623 TN Soulsville Charter School 38106
#> 17624 TN KIPP Memphis Collegiate High School 381083.3 Creating variables
3.3.1 Create new data frame based on df_school_all
Data frame df_school_all has one obs per US high school and then variables identifying number of visits by particular universities
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_school_allvars.RData"))
names(df_school_all)
#> [1] "state_code" "school_type" "ncessch"
#> [4] "name" "address" "city"
#> [7] "zip_code" "pct_white" "pct_black"
#> [10] "pct_hispanic" "pct_asian" "pct_amerindian"
#> [13] "pct_other" "num_fr_lunch" "total_students"
#> [16] "num_took_math" "num_prof_math" "num_took_rla"
#> [19] "num_prof_rla" "avgmedian_inc_2564" "latitude"
#> [22] "longitude" "visits_by_196097" "visits_by_186380"
#> [25] "visits_by_215293" "visits_by_201885" "visits_by_181464"
#> [28] "visits_by_139959" "visits_by_218663" "visits_by_100751"
#> [31] "visits_by_199193" "visits_by_110635" "visits_by_110653"
#> [34] "visits_by_126614" "visits_by_155317" "visits_by_106397"
#> [37] "visits_by_149222" "visits_by_166629" "total_visits"
#> [40] "inst_196097" "inst_186380" "inst_215293"
#> [43] "inst_201885" "inst_181464" "inst_139959"
#> [46] "inst_218663" "inst_100751" "inst_199193"
#> [49] "inst_110635" "inst_110653" "inst_126614"
#> [52] "inst_155317" "inst_106397" "inst_149222"
#> [55] "inst_166629"Create new version of data frame, called school_v2, which we’ll use to introduce how to create new variables
library(tidyverse) # below code use tidyverse functions and pipe operator
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ ggplot2 3.4.3 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
school_v2 <- df_school_all %>%
select(-contains("inst_")) %>% # remove vars that start with "inst_"
rename( # rename selected variables
visits_by_berkeley = visits_by_110635,
visits_by_boulder = visits_by_126614,
visits_by_bama = visits_by_100751,
visits_by_stonybrook = visits_by_196097,
visits_by_rutgers = visits_by_186380,
visits_by_pitt = visits_by_215293,
visits_by_cinci = visits_by_201885,
visits_by_nebraska = visits_by_181464,
visits_by_georgia = visits_by_139959,
visits_by_scarolina = visits_by_218663,
visits_by_ncstate = visits_by_199193,
visits_by_irvine = visits_by_110653,
visits_by_kansas = visits_by_155317,
visits_by_arkansas = visits_by_106397,
visits_by_sillinois = visits_by_149222,
visits_by_umass = visits_by_166629,
num_took_read = num_took_rla,
num_prof_read = num_prof_rla,
med_inc = avgmedian_inc_2564
)
glimpse(school_v2)3.3.2 Base R approach to creating new variables
Create new variables using assignment operator <- and subsetting operators [] and $ to create new variables and set conditions of the input variables
Pseudo syntax: df$newvar <- ...
- where
...argument is expression(s)/calculation(s) used to create new variables- expressions can include subsetting operators and/or other base R functions
Task: Create measure of percent of students on free-reduced lunch
base R approach
school_v2_temp<- school_v2 #create copy of dataset; not necessary
school_v2_temp$pct_fr_lunch <-
school_v2_temp$num_fr_lunch/school_v2_temp$total_students
#investigate variable you created
str(school_v2_temp$pct_fr_lunch)
#> num [1:21301] 0.723 1 0.967 0.93 1 ...
school_v2_temp$pct_fr_lunch[1:5] # print first 5 obs
#> [1] 0.7225549 1.0000000 0.9666667 0.9303483 1.0000000tidyverse approach (with pipes)
school_v2_temp <- school_v2 %>%
mutate(pct_fr_lunch = num_fr_lunch/total_students) If creating new variable based on the condition/values of input variables, basically the tidyverse equivalent of mutate() with if_else() or recode()
- Pseudo syntax:
df$newvar[logical condition]<- new value logical condition: a condition that evaluates toTRUEorFALSE
Task: Create 0/1 indicator if school has median income greater than $100k
tidyverse approach (using pipes)
school_v2_temp %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0)) %>%
count(inc_gt_100k) # note how NA values of med_inc treated
#> # A tibble: 3 × 2
#> inc_gt_100k n
#> <dbl> <int>
#> 1 0 18632
#> 2 1 2045
#> 3 NA 624Base R approach
school_v2_temp$inc_gt_100k<-NA #initialize an empty column with NAs
# otherwise you'll get warning
school_v2_temp$inc_gt_100k[school_v2_temp$med_inc>100000] <- 1
school_v2_temp$inc_gt_100k[school_v2_temp$med_inc<=100000] <- 0
count(school_v2_temp, inc_gt_100k)
#> # A tibble: 3 × 2
#> inc_gt_100k n
#> <dbl> <int>
#> 1 0 18632
#> 2 1 2045
#> 3 NA 624Task: Using data frame wwlist and input vars state and firstgen, create a 4-category var with following categories:
- “instate_firstgen”; “instate_nonfirstgen”; “outstate_firstgen”; “outstate_nonfirstgen”
tidyverse approach (using pipes)
load(url("https://github.com/ozanj/rclass/raw/master/data/prospect_list/wwlist_merged.RData"))
wwlist_temp <- wwlist %>%
mutate(state_gen = case_when(
state == "WA" & firstgen =="Y" ~ "instate_firstgen",
state == "WA" & firstgen =="N" ~ "instate_nonfirstgen",
state != "WA" & firstgen =="Y" ~ "outstate_firstgen",
state != "WA" & firstgen =="N" ~ "outstate_nonfirstgen")
)
str(wwlist_temp$state_gen)
#> chr [1:268396] NA "instate_nonfirstgen" "instate_nonfirstgen" ...
wwlist_temp %>% count(state_gen)
#> # A tibble: 5 × 2
#> state_gen n
#> <chr> <int>
#> 1 instate_firstgen 32428
#> 2 instate_nonfirstgen 58646
#> 3 outstate_firstgen 32606
#> 4 outstate_nonfirstgen 134616
#> 5 <NA> 10100Task: Using wwlist and input vars state and firstgen, create a 4-category var
base R approach
wwlist_temp <- wwlist
wwlist_temp$state_gen <- NA
wwlist_temp$state_gen[wwlist_temp$state == "WA"
& wwlist_temp$firstgen =="Y"] <- "instate_firstgen"
wwlist_temp$state_gen[wwlist_temp$state == "WA"
& wwlist_temp$firstgen =="N"] <- "instate_nonfirstgen"
wwlist_temp$state_gen[wwlist_temp$state != "WA"
& wwlist_temp$firstgen =="Y"] <- "outstate_firstgen"
wwlist_temp$state_gen[wwlist_temp$state != "WA"
& wwlist_temp$firstgen =="N"] <- "outstate_nonfirstgen"
str(wwlist_temp$state_gen)
#> chr [1:268396] NA "instate_nonfirstgen" "instate_nonfirstgen" ...
count(wwlist_temp, state_gen)
#> # A tibble: 5 × 2
#> state_gen n
#> <chr> <int>
#> 1 instate_firstgen 32428
#> 2 instate_nonfirstgen 58646
#> 3 outstate_firstgen 32606
#> 4 outstate_nonfirstgen 134616
#> 5 <NA> 101003.4 Sorting data
3.4.1 Base R sort() for vectors
sort() is a base R function that sorts vectors
Syntax: sort(x, decreasing=FALSE, ...)
- where x is object being sorted
- By default it sorts in ascending order (low to high)
- Need to set decreasing argument to
TRUEto sort from high to low
#?sort()
x<- c(31, 5, 8, 2, 25)
sort(x)
#> [1] 2 5 8 25 31
sort(x, decreasing = TRUE)
#> [1] 31 25 8 5 23.4.2 Base R order() for dataframes
order() is a base R function that sorts vectors
- Syntax:
order(..., na.last = TRUE, decreasing = FALSE) - where
...are variable(s) to sort by - By default it sorts in ascending order (low to high)
- Need to set decreasing argument to
TRUEto sort from high to low
Descending argument only works when we want either one (and only) variable descending or all variables descending (when sorting by multiple vars)
- use
-when you want to indicate which variables are descending while using the default ascending sorting
df_event[order(df_event$event_date), ]
df_event[order(df_event$event_date, df_event$total_12), ]
#sort descending via argument
df_event[order(df_event$event_date, decreasing = TRUE), ]
df_event[order(df_event$event_date, df_event$total_12, decreasing = TRUE), ]
#sorting by both ascending and descending variables
df_event[order(df_event$event_date, -df_event$total_12), ]3.4.3 Example, sorting
- Create a new dataframe from df_events that sorts by ascending by
event_date, ascendingevent_state, and descendingpop_total.
base R using order() function:
df_event_br1 <- df_event[order(df_event$event_date, df_event$event_state,
-df_event$pop_total), ]