library(tidyverse)Into the tidyverse: dplyr, select, filter, and pipes
1 Introduction
1.1 Libraries we will use today
“Load” the package we will use today (output omitted)
- you must run this code chunk
If package not yet installed, then must install before you load. Install in “console” rather than .qmd file
- Generic syntax:
install.packages("package_name") - Install “tidyverse”:
install.packages("tidyverse")
Note: when we load package, name of package is not in quotes; but when we install package, name of package is in quotes:
install.packages("tidyverse")library(tidyverse)
1.2 Data for sections on select(), filter(), and arrange() functions
Load .Rdata data frames, df_event and df_school
Data on off-campus recruiting events by public universities
- Data frame object
df_event- One observation per university, recruiting event
- Data frame object
df_school- One observation per high school (visited and non-visited)
rm(list = ls()) # remove all objects in current environment
getwd()
#> [1] "/Users/jaquette/Documents/rclass1/lectures/into_the_tidyverse"
#load dataset with one obs per recruiting event
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_event_somevars.RData"))
#load("../../data/recruiting/recruit_event_somevars.Rdata")
#load dataset with one obs per high school
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_school_somevars.RData"))
#load("../../data/recruiting/recruit_school_somevars.Rdata")1.3 Data for sections on pipes %>% and mutate() function
Load .Rdata data frame wwlist, “prospects” purchased by Western Washington U.
Note: we won’t use this data frame until the lecture section on “pipes”
- You can ignore
wwlistdata frame for lecture sections on select(), filter(), and arrange() functions
The “Student list” business
- Universities identify/target “prospects” by buying “student lists” from College Board/ACT (e.g., $.50+ per prospect)
- Prospect lists contain contact info (e.g., address, email), academic achievement, socioeconomic, demographic characteristics
- Universities choose which prospects to purchase by filtering on criteria like zip-code, GPA, test score range, etc.
#load prospect list data
load(url("https://github.com/ozanj/rclass/raw/master/data/prospect_list/wwlist_merged.RData"))Object wwlist
- De-identified list of prospective students purchased by Western Washington University from College Board
- We collected these data using public records requests request
1.3.1 Data frame wwlist, “prospects” purchased by Western Washington U.
Observations on wwlist
- each observation represents a prospective student
typeof(wwlist)
#> [1] "list"
dim(wwlist)
#> [1] 268396 41Variables on wwlist
some vars provide de-identified data on individual prospects
- e.g.,
psat_range,state,sex,ethn_code
- e.g.,
some vars provide data about zip-code student lives in
- e.g.,
med_inc,pop_total,pop_black
- e.g.,
some vars provide data about school student enrolled in
- e.g.,
fr_lunchis number of students on free/reduced lunch - note: bad merge between prospect-level data and school-level data
- e.g.,
names(wwlist)
str(wwlist)
glimpse(wwlist) # tidyverse function, similar to str()Variable firstgen identifies whether prospect is a first-generation college student
Imagine we want to isolate all the first-generation prospects
- Investigate variable type/structure.
- A dichotomous var, but stored as character in
wwlist. So must use quotes (''or"") to filter/subset based on values offirstgen
str(wwlist$firstgen)
#> chr [1:268396] NA "N" "N" "N" NA "N" "N" "Y" "Y" "N" "N" "N" "N" "N" "N" ...- Create frequency table to identify possible values of
firstgen
table(wwlist$firstgen, useNA = "always")
#>
#> N Y <NA>
#> 193333 65046 10017- Isolate all the first-gen prospects (output omitted)
filter(wwlist, firstgen == "Y")2 Investigating data patterns
2.0.1 Introduction to the dplyr library
dplyr, a package within the tidyverse suite of packages, provide tools for manipulating data frames
- Wickham describes functions within
dplyras a set of “verbs” that fall in the broader categories of subsetting, sorting, and transforming
| Today | Upcoming weeks |
|---|---|
| Subsetting data | Transforming data |
- select() variables |
- mutate() creates new variables |
- filter() observations |
- summarize() calculates across rows |
| Sorting data | - group_by() to calculate across rows within groups |
- arrange() |
All dplyr verbs (i.e., functions) work as follows
- first argument is a data frame
- subsequent arguments describe what to do with variables and observations in data frame
- refer to variable names without quotes
- result of the function is a new data frame
2.1 select() variables
Select variables using select() function
Printing observations is key to investigating data, but datasets often have hundreds, thousands of variables
select() function selects columns of data (i.e., variables) you specify
- first argument is the name of data frame object
- remaining arguments are variable names, which are separated by commas and without quotes
Without assignment (<-), select() by itself simply prints selected vars
#?select
select(df_event,instnm,event_date,event_type,event_state,med_inc)
#> # A tibble: 18,680 × 5
#> instnm event_date event_type event_state med_inc
#> <chr> <date> <chr> <chr> <dbl>
#> 1 UM Amherst 2017-10-12 public hs MA 71714.
#> 2 UM Amherst 2017-10-04 public hs MA 89122.
#> 3 UM Amherst 2017-10-25 public hs MA 70136.
#> 4 UM Amherst 2017-10-26 public hs MA 70136.
#> 5 Stony Brook 2017-10-02 public hs MA 71024.
#> 6 USCC 2017-09-18 private hs MA 71024.
#> 7 UM Amherst 2017-09-18 private hs MA 71024.
#> 8 UM Amherst 2017-09-26 public hs MA 97225
#> 9 UM Amherst 2017-09-26 private hs MA 97225
#> 10 UM Amherst 2017-10-12 public hs MA 77800.
#> # ℹ 18,670 more rowsRecall that all dplyr functions (e.g., select()) return a new data frame object
- type equals “list”
- length equals number of vars you select
typeof(select(df_event,instnm,event_date,event_type,event_state,med_inc))
#> [1] "list"
length(select(df_event,instnm,event_date,event_type,event_state,med_inc))
#> [1] 5glimpse(): tidyverse function for viewing data frames
- a cross between
str()and simply printing data
?glimpse
glimpse(df_event)glimpse() a select() set of variables
glimpse(select(df_event,instnm,event_date,event_type,event_state,med_inc))
#> Rows: 18,680
#> Columns: 5
#> $ instnm <chr> "UM Amherst", "UM Amherst", "UM Amherst", "UM Amherst", "S…
#> $ event_date <date> 2017-10-12, 2017-10-04, 2017-10-25, 2017-10-26, 2017-10-0…
#> $ event_type <chr> "public hs", "public hs", "public hs", "public hs", "publi…
#> $ event_state <chr> "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA"…
#> $ med_inc <dbl> 71713.5, 89121.5, 70136.5, 70136.5, 71023.5, 71023.5, 7102…With assignment (<-), select() creates a new object containing only the variables you specify
event_small <- select(df_event,instnm,event_date,event_type,event_state,
med_inc)
glimpse(event_small)
#> Rows: 18,680
#> Columns: 5
#> $ instnm <chr> "UM Amherst", "UM Amherst", "UM Amherst", "UM Amherst", "S…
#> $ event_date <date> 2017-10-12, 2017-10-04, 2017-10-25, 2017-10-26, 2017-10-0…
#> $ event_type <chr> "public hs", "public hs", "public hs", "public hs", "publi…
#> $ event_state <chr> "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA", "MA"…
#> $ med_inc <dbl> 71713.5, 89121.5, 70136.5, 70136.5, 71023.5, 71023.5, 7102…Select helper functions
select() can use “helper functions” starts_with(), contains(), and ends_with() to choose columns
?selectExample:
#names(df_event)
select(df_event,instnm,starts_with("event"))
#> # A tibble: 18,680 × 8
#> instnm event_date event_type event_state event_inst event_name
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 UM Amherst 2017-10-12 public hs MA In-State Amherst-Pelham Regi…
#> 2 UM Amherst 2017-10-04 public hs MA In-State Hampshire County Co…
#> 3 UM Amherst 2017-10-25 public hs MA In-State Chicopee High Schoo…
#> 4 UM Amherst 2017-10-26 public hs MA In-State Chicopee Comprehens…
#> 5 Stony Brook 2017-10-02 public hs MA Out-State Easthampton High Sc…
#> 6 USCC 2017-09-18 private hs MA Out-State Williston Northampt…
#> 7 UM Amherst 2017-09-18 private hs MA In-State Williston-Northampt…
#> 8 UM Amherst 2017-09-26 public hs MA In-State Granby Jr Sr High S…
#> 9 UM Amherst 2017-09-26 private hs MA In-State MacDuffie School Vi…
#> 10 UM Amherst 2017-10-12 public hs MA In-State Smith Academy Visit
#> # ℹ 18,670 more rows
#> # ℹ 2 more variables: event_location_name <chr>, event_datetime_start <dttm>2.2 Rename variables
rename() function renames variables within a data frame object
Syntax:
rename(obj_name, new_name = old_name,...)
rename(df_event, g12_offered = g12offered,
titlei = titlei_status_pub)
names(df_event)Variable names do not change permanently unless we combine rename with assignment
rename_event <- rename(df_event, g12_offered = g12offered, titlei = titlei_status_pub)
names(rename_event)
rm(rename_event)2.3 count() function
count() function from dplyr package counts the unique values of one or more variables.
Syntax [see help file for full syntax]
count(x,...)
Arguments [see help file for full arguments]
x: an object, often a data frame...: variables to group by
Examples of using count()
- Without vars in
...argument, counts number of obs in object
count(df_school)
# df_school %>% count() # same as above but using pipes
str(count(df_school))
# #df_school %>% count() %>% str() # same as above but using pipesWith vars in
...argument, counts number of obs per variable value- This is the best way to create frequency table, better than
table() - note: by default,
count()always showsNAs[this is good!]
count(df_school,school_type) # df_school %>% count(school_type) # same as above but using pipes str(count(df_school,school_type)) # df_school %>% count(school_type) %>% str() # same as above but using pipes- This is the best way to create frequency table, better than
2.4 filter() rows
The filter() function
filter() allows you to select observations based on values of variables
- Arguments
- first argument is name of data frame
- subsequent arguments are logical expressions to filter the data frame
- Multiple expressions separated by commas work as AND operators (e.g., condtion 1
TRUEAND condition 2TRUE)
- What is the result of a
filter()command?filter()returns a data frame consisting of rows where the condition isTRUE
?filterExample from data frame object df_school, each obs is a high school
- Show all obs where the high school received 1 visit from UC Berkeley (110635) [output omitted]
filter(df_school,visits_by_110635 == 1)Note that resulting object is list, consisting of obs where condition TRUE
nrow(df_school)
#> [1] 21301
nrow(filter(df_school,visits_by_110635 == 1))
#> [1] 528The filter() function
Task: Count the number of high schools that received 1 visit from UC Berkeley.
[dplyr] Using filter():
nrow(filter(df_school, visits_by_110635 == 1))
#> [1] 528Filter, character variables
Use single quotes '' or double quotes "" to refer to values of character variables
glimpse(select(df_school, school_type, state_code))
#> Rows: 21,301
#> Columns: 2
#> $ school_type <chr> "public", "public", "public", "public", "public", "public"…
#> $ state_code <chr> "AK", "AK", "AK", "AK", "AK", "AK", "AK", "AK", "AK", "AK"…Identify all private high schools in CA that got 1 visit by particular universities
- Visited once by UC Berkeley (ID=110635)
filter(df_school,visits_by_110635 == 1, school_type == "private",
state_code == "CA")- Visited once by University of Alabama (ID=100751)
filter(df_school,visits_by_100751 == 1, school_type == "private",
state_code == "CA") - Visited once by Berkeley and University of Alabama
filter(df_school,visits_by_100751 == 1, visits_by_110635 == 1,
school_type == "private", state_code == "CA") Filter by multiple conditions
Task: Count the number of private high schools in CA that received 1 visit each from UC Berkeley and University of Alabama.
Using filter():
nrow(filter(df_school, visits_by_100751 == 1, visits_by_110635 == 1,
school_type == "private", state_code == "CA"))
#> [1] 92.4.1 Logical operators for comparisons
logical operators useful for: filter obs w/ filter(); create variables w/ mutate()
- logical operators also work when using Base R functions
| Operator symbol | Operator meaning |
|---|---|
== |
Equal to |
!= |
Not equal to |
> |
greater than |
>= |
greater than or equal to |
< |
less than |
<= |
less than or equal to |
& |
AND |
| |
OR |
%in% |
includes |
- Visualization of “Boolean” operators (e.g., AND, OR, AND NOT)
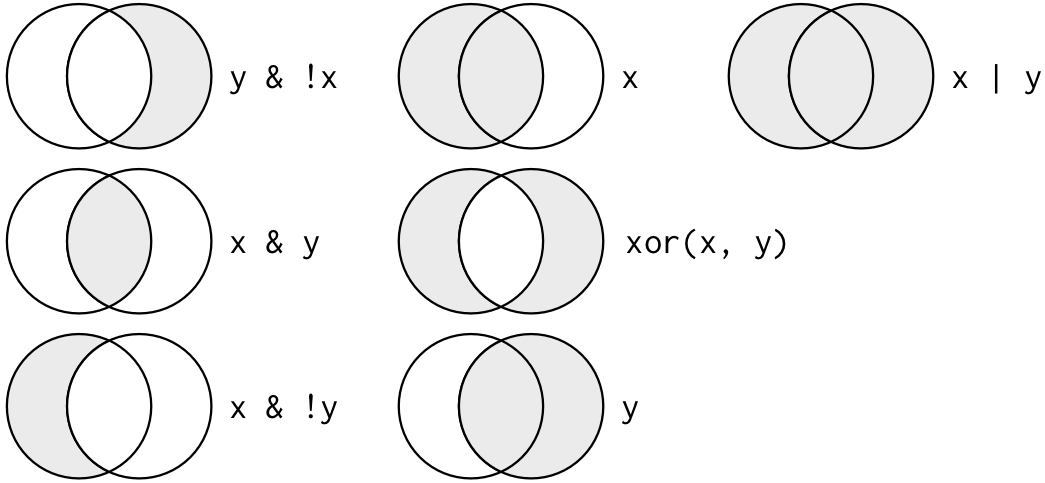
2.4.2 Filters and comparisons, Demonstration
Schools visited by Bama (100751) and/or Berkeley (110635)
# Berkeley AND Bama
filter(df_school,visits_by_100751 >= 1, visits_by_110635 >= 1)
filter(df_school,visits_by_100751 >= 1 & visits_by_110635 >= 1) # same same# Berkeley OR Bama
filter(df_school,visits_by_100751 >= 1 | visits_by_110635 >= 1)Apply count() function on top of filter() function to count the number of observations that satisfy criteria
- Avoids printing individual observations
# Number of schools that get visit by Berkeley AND Bama
count(filter(df_school, visits_by_100751 >= 1 & visits_by_110635 >= 1))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 247
# Number of schools that get visit by Berkeley OR Bama
count(filter(df_school, visits_by_100751 >= 1 | visits_by_110635 >= 1))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 2763- Note: You could also use any of the base R equivalents from the Base R lecture
2.4.3 Filters and comparisons, >=
Number of public high schools that are at least 50% Black in Alabama compared to number of schools that received visit by Bama:
# at least 50% black
count(filter(df_school, school_type == "public", pct_black >= 50,
state_code == "AL"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 86
# at least 50% black and received visit by Bama
count(filter(df_school, school_type == "public", pct_black >= 50,
state_code == "AL", visits_by_100751 >= 1))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 21Number of public high schools that are at least 50% White in Alabama compared to number of schools that received visit by Bama:
# at least 50% white
count(filter(df_school, school_type == "public", pct_white >= 50,
state_code == "AL"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 238
# at least 50% white and received visit by Bama
count(filter(df_school, school_type == "public", pct_white >= 50,
state_code == "AL", visits_by_100751 >= 1))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 822.4.4 Filters and comparisons, not equals (!=)
Count the number of high schools visited by University of Colorado (126614) that are not located in CO
#number of high schools visited by U Colorado
count(filter(df_school, visits_by_126614 >= 1))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1056
#number of high schools visited by U Colorado not located in CO
count(filter(df_school, visits_by_126614 >= 1, state_code != "CO"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 873
#number of high schools visited by U Colorado located in CO
#count(filter(df_school, visits_by_126614 >= 1, state_code == "CO"))2.4.5 Filters and comparisons, %in% operator
What if you wanted to count the number of schools visited by Bama (100751) in a group of states?
count(filter(df_school,visits_by_100751 >= 1, state_code == "MA" |
state_code == "VT" | state_code == "ME"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 108Easier way to do this is with %in% operator
count(filter(df_school,visits_by_100751 >= 1, state_code %in% c("MA","ME","VT")))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 108Select the private high schools that got either 2 or 3 visits from Bama
count(filter(df_school, visits_by_100751 %in% 2:3, school_type == "private"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1832.4.6 Identifying data type and possible values helpful for filtering
typeof()andstr()shows internal data type of a variabletable()to show potential values of categorical variables
typeof(df_event$event_type)
#> [1] "character"
str(df_event$event_type) # double quotes indicate character
#> chr [1:18680] "public hs" "public hs" "public hs" "public hs" "public hs" ...
table(df_event$event_type, useNA="always")
#>
#> 2yr college 4yr college other private hs public hs <NA>
#> 951 531 2001 3774 11423 0
typeof(df_event$med_inc)
#> [1] "double"
str(df_event$med_inc)
#> num [1:18680] 71714 89122 70136 70136 71024 ...Now that we know event_type is a character, we can filter values
count(filter(df_event, event_type == "public hs", event_state =="CA"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1100
#below code would return an error because variables are character
#count(filter(df_event, event_type == public hs, event_state ==CA))2.4.7 Filtering and missing values
Wickham (2018) states:
- “
filter()only includes rows where condition is TRUE; it excludes bothFALSEandNAvalues. To preserve missing values, ask for them explicitly:”
Investigate var df_event$fr_lunch, number of free/reduced lunch students
- only available for visits to public high schools
#visits to public HS with less than 50 students on free/reduced lunch
count(filter(df_event,event_type == "public hs", fr_lunch<50))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 910
#visits to public HS, where free/reduced lunch missing
count(filter(df_event,event_type == "public hs", is.na(fr_lunch)))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 26
#visits to public HS, where free/reduced is less than 50 OR is missing
count(filter(df_event,event_type == "public hs", fr_lunch<50 | is.na(fr_lunch)))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 9362.4.8 Exercises
Use the data from df_event, which has one observation for each off-campus recruiting event a university attends
- Count the number of events attended by the University of Pittsburgh (Pitt)
univ_id == 215293
- Count the number of recruiting events by Pitt at public or private high schools
- Count the number of recruiting events by Pitt at public or private high schools located in the state of PA
- Count the number of recruiting events by Pitt at public high schools not located in PA where median income is less than 100,000
- Count the number of recruiting events by Pitt at public high schools not located in PA where median income is greater than or equal to 100,000
- Count the number of out-of-state recruiting events by Pitt at private high schools or public high schools with median income of at least 100,000
Solutions
- Count the number of events attended by the University of Pittsburgh (Pitt)
univ_id == 215293
count(filter(df_event, univ_id == 215293))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1225- Count the number of recruiting events by Pitt at public or private high schools
str(df_event$event_type)
#> chr [1:18680] "public hs" "public hs" "public hs" "public hs" "public hs" ...
table(df_event$event_type, useNA = "always")
#>
#> 2yr college 4yr college other private hs public hs <NA>
#> 951 531 2001 3774 11423 0
count(filter(df_event, univ_id == 215293, event_type == "private hs" |
event_type == "public hs"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1030- Count the number of recruiting events by Pitt at public or private high schools located in the state of PA
count(filter(df_event, univ_id == 215293, event_type == "private hs" |
event_type == "public hs", event_state == "PA"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 262- Count the number of recruiting events by Pitt at public high schools not located in PA where median income is less than 100,000
count(filter(df_event, univ_id == 215293, event_type == "public hs",
event_state != "PA", med_inc < 100000))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 213- Count the number of recruiting events by Pitt at public high schools not located in PA where median income is greater than or equal to 100,000
count(filter(df_event, univ_id == 215293, event_type == "public hs",
event_state != "PA", med_inc >= 100000))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 344- Count the number of out-of-state recruiting events by Pitt at private high schools or public high schools with median income of at least 100,000
count(filter(df_event, univ_id == 215293, event_state != "PA",
(event_type == "public hs" & med_inc >= 100000) |
event_type == "private hs"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 5532.5 arrange() rows (i.e., sort rows)
2.5.1 arrange() function
arrange() function “arranges” rows in a data frame; said different, it sorts observations
Syntax: arrange(x,...)
- First argument,
x, is a data frame - Subsequent arguments are a “comma separated list of unquoted variable names”
df_event
arrange(df_event, event_date)Data frame goes back to previous order unless you assign the new order
df_event
df_event <- arrange(df_event, event_date)
df_eventAscending and descending order
arrange()sorts in ascending order by default- use
desc()to sort a column by descending order
arrange(df_event, desc(event_date))Can sort by multiple variables
arrange(df_event, univ_id, desc(event_date), desc(med_inc))
#sort by university and descending by size of 12th grade class; combine with select
select(arrange(df_event, univ_id, desc(g12)),instnm,event_type,event_date,g12)2.5.2 arrange(), missing values sorted at the end
Missing values automatically sorted at the end, regardless of whether you sort ascending or descending
Below, we sort by university, then by date of event, then by ID of high school
#by university, date, ascending school id
select(arrange(df_event, univ_id, desc(event_date), school_id),
instnm,event_date,event_type,school_id)
#by university, date, descending school id
select(arrange(df_event, univ_id, desc(event_date), desc(school_id)),
instnm,event_date,event_type,school_id)Can sort by is.na to put missing values first
select(arrange(df_event, univ_id, desc(event_date), desc(is.na(school_id))),
instnm,event_date,event_type,school_id)
#> # A tibble: 18,680 × 4
#> instnm event_date event_type school_id
#> <chr> <date> <chr> <chr>
#> 1 Bama 2017-12-18 other <NA>
#> 2 Bama 2017-12-18 private hs A9106483
#> 3 Bama 2017-12-15 other <NA>
#> 4 Bama 2017-12-15 public hs 484473005095
#> 5 Bama 2017-12-15 public hs 062927004516
#> 6 Bama 2017-12-14 other <NA>
#> 7 Bama 2017-12-13 other <NA>
#> 8 Bama 2017-12-13 public hs 130387001439
#> 9 Bama 2017-12-13 private hs 00071151
#> 10 Bama 2017-12-13 public hs 063386005296
#> # ℹ 18,670 more rows2.5.3 Exercise, arranging
Use the data from df_event, which has one observation for each off-campus recruiting event a university attends
- Sort ascending by “univ_id” and descending by “event_date”
- Select four variables in total and sort ascending by “univ_id” and descending by “event_date”
- Now using the same variables from above, sort by
is.nato put missing values in “school_id” first
Solutions
- Sort ascending by “univ_id” and descending by “event_date”
arrange(df_event, univ_id, desc(event_date))
#> # A tibble: 18,680 × 33
#> instnm univ_id instst pid event_date event_type zip school_id ipeds_id
#> <chr> <int> <chr> <int> <date> <chr> <chr> <chr> <int>
#> 1 Bama 100751 AL 7115 2017-12-18 private hs 77089 A9106483 NA
#> 2 Bama 100751 AL 7121 2017-12-18 other <NA> <NA> NA
#> 3 Bama 100751 AL 7114 2017-12-15 public hs 75165 484473005095 NA
#> 4 Bama 100751 AL 7100 2017-12-15 public hs 93012 062927004516 NA
#> 5 Bama 100751 AL 7073 2017-12-15 other 98027 <NA> NA
#> 6 Bama 100751 AL 7072 2017-12-14 other 98007 <NA> NA
#> 7 Bama 100751 AL 7118 2017-12-13 public hs 31906 130387001439 NA
#> 8 Bama 100751 AL 7099 2017-12-13 private hs 90293 00071151 NA
#> 9 Bama 100751 AL 7109 2017-12-13 public hs 92630 063386005296 NA
#> 10 Bama 100751 AL 7071 2017-12-13 other 98032 <NA> NA
#> # ℹ 18,670 more rows
#> # ℹ 24 more variables: event_state <chr>, event_inst <chr>, med_inc <dbl>,
#> # pop_total <dbl>, pct_white_zip <dbl>, pct_black_zip <dbl>,
#> # pct_asian_zip <dbl>, pct_hispanic_zip <dbl>, pct_amerindian_zip <dbl>,
#> # pct_nativehawaii_zip <dbl>, pct_tworaces_zip <dbl>,
#> # pct_otherrace_zip <dbl>, fr_lunch <dbl>, titlei_status_pub <fct>,
#> # total_12 <dbl>, school_type_pri <int>, school_type_pub <int>, …- Select four variables in total and sort ascending by “univ_id” and descending by “event_date”
select(arrange(df_event, univ_id, desc(event_date)), univ_id, event_date,
instnm, event_type)
#> # A tibble: 18,680 × 4
#> univ_id event_date instnm event_type
#> <int> <date> <chr> <chr>
#> 1 100751 2017-12-18 Bama private hs
#> 2 100751 2017-12-18 Bama other
#> 3 100751 2017-12-15 Bama public hs
#> 4 100751 2017-12-15 Bama public hs
#> 5 100751 2017-12-15 Bama other
#> 6 100751 2017-12-14 Bama other
#> 7 100751 2017-12-13 Bama public hs
#> 8 100751 2017-12-13 Bama private hs
#> 9 100751 2017-12-13 Bama public hs
#> 10 100751 2017-12-13 Bama other
#> # ℹ 18,670 more rows- Select the variables “univ_id”, “event_date”, and “school_id” and sort by
is.nato put missing values in “school_id” first.
select(arrange(df_event, univ_id, desc(event_date), desc(is.na(school_id))),
univ_id, event_date, school_id)
#> # A tibble: 18,680 × 3
#> univ_id event_date school_id
#> <int> <date> <chr>
#> 1 100751 2017-12-18 <NA>
#> 2 100751 2017-12-18 A9106483
#> 3 100751 2017-12-15 <NA>
#> 4 100751 2017-12-15 484473005095
#> 5 100751 2017-12-15 062927004516
#> 6 100751 2017-12-14 <NA>
#> 7 100751 2017-12-13 <NA>
#> 8 100751 2017-12-13 130387001439
#> 9 100751 2017-12-13 00071151
#> 10 100751 2017-12-13 063386005296
#> # ℹ 18,670 more rows3 Pipes
3.1 What are “pipes”, %>%
Pipes are a means of performing multiple steps in a single line of code
- When writing code, the pipe symbol is
%>% - The pipe operator
%>%is created by the magrittr package, which is not part of base R - However, the magrittr package is automatically loaded when you load the tidyverse package
?magrittr::`%>%`pipe syntax: LHS %>% RHS
LHS(refers to “left hand side” of the pipe) is an object or functionRHS(refers to “right hand side” of the pipe) is a function
How pipes work:
- Object created by
LHSbecomes the first argument of the function (RHS) to the right of the%>%pipe symbol - Basic code flow:
object %>% function1 %>% function2 %>% function3
- Output of
some_function1becomes the input (the first argument) of the functionsome_function2to the right of the%>%pipe symbol
Example of using pipes to calculate mean value of atomic vector
1:10 # an atomic vector
#> [1] 1 2 3 4 5 6 7 8 9 10
mean(1:10) # calculate mean without pipes
#> [1] 5.5
1:10 %>% mean() # calculate mean with pipes
#> [1] 5.5- no pipe: (1) write function; (2) data object
1:10is 1st argument ofmean() - pipe: (1) write data object; (2) “pipe” (verb) object as 1st argument of
mean()
Intuitive mnemonic device for understanding pipes
- whenever you see a pipe
%>%think of the words “and then…”
Example: isolate all the first-generation prospects [output omitted]
- in words: start with object
wwlistand then filter first generation students
wwlist %>% filter(firstgen == "Y")below code in words:
- start with
wwlistand then select a few vars and then filter and then sort and then investigate the structure of object
wwlist %>% select(firstgen, state, med_inc_zip) %>%
filter(firstgen == "Y", state == "WA") %>%
arrange(desc(med_inc_zip)) %>% str()
#> tibble [32,428 × 3] (S3: tbl_df/tbl/data.frame)
#> $ firstgen : chr [1:32428] "Y" "Y" "Y" "Y" ...
#> $ state : chr [1:32428] "WA" "WA" "WA" "WA" ...
#> $ med_inc_zip: num [1:32428] 216720 216720 216720 216720 216720 ...Example: apply “structure” function str() to wwlist with and without pipes
str(wwlist) # without pipe
wwlist %>% str() # with pipeI use the str() when I add new %>%; shows what kind of object being piped in
- task: select a few vars from
wwlist; isolate first-gen students in WA; sort descending by income (output omitted)
wwlist %>% select(firstgen, state, med_inc_zip) %>% str()
wwlist %>% select(firstgen, state, med_inc_zip) %>%
filter(firstgen == "Y", state == "WA") %>% str()
wwlist %>% select(firstgen, state, med_inc_zip) %>%
filter(firstgen == "Y", state == "WA") %>%
arrange(desc(med_inc_zip)) %>% str()3.2 Compare data tasks, with and without pipes
Task: Using object wwlist print data for “first-gen” prospects (firstgen == "Y")
# without pipes
filter(wwlist, firstgen == "Y")
# with pipes
wwlist %>% filter(firstgen == "Y")Comparing the two approaches:
- “without pipes”, object
wwlistis the first argumentfilter()function - In “pipes” approach, you don’t specify object
wwlistas first argument infilter()- Why? Because
%>%“pipes” the object to the left of the%>%operator into the function to the right of the%>%operator
- Why? Because
Task: Using object wwlist, print data for “first-gen” prospects for selected variables
#Without pipes
select(filter(wwlist, firstgen == "Y"), state, hs_city, sex)
#With pipes
wwlist %>% filter(firstgen == "Y") %>% select(state, hs_city, sex)Comparing the two approaches:
- In the “without pipes” approach, code is written “inside out”
- The first step in the task – identifying the object – is the innermost part of code
- The last step in task – selecting variables to print – is the outermost part of code
- In “pipes” approach the left-to-right order of code matches how we think about the task
- First, we start with an object and then (
%>%) we usefilter()to isolate first-gen students and then (%>%) we select which variables to print
- First, we start with an object and then (
str() helpful to understand object piped in from one function to another
#object that was "piped" into `select()` from `filter()`
wwlist %>% filter(firstgen == "Y") %>% str()
#object that was created after `select()` function
wwlist %>% filter(firstgen == "Y") %>% select(state, hs_city, sex) %>% str()3.3 count() function with pipes %>%
Recall count() function from dplyr package counts the number of obs by group
Syntax [see help file for full syntax]
count(x,...)
Arguments [see help file for full arguments]
x: an object, often a data frame...: variables to group by
Examples of using count()
- Without vars in
...argument, counts number of obs in object
count(wwlist)
wwlist %>% count()
wwlist %>% count() %>% str()With vars in
...argument, counts number of obs per variable value- This is the best way to create frequency table, better than
table() - note: by default,
count()always showsNAs[this is good!]
- This is the best way to create frequency table, better than
count(wwlist,school_category)
wwlist %>% count(school_category)
wwlist %>% count(school_category) %>% str()3.4 pipe operators and new lines
Often want to insert line breaks to make long line of code more readable
- When inserting line breaks, pipe operator
%>%should be the last thing before a line break, not the first thing after a line break
This works
wwlist %>% filter(firstgen == "Y") %>%
select(state, hs_city, sex) %>%
count(sex)This works too
wwlist %>% filter(firstgen == "Y",
state != "WA") %>%
select(state, hs_city, sex) %>%
count(sex)This doesn’t work
wwlist %>% filter(firstgen == "Y")
%>% select(state, hs_city, sex)
%>% count(sex)3.5 The power of pipes
You might be thinking, “what’s the big deal?”
Task:
- in one line of code, modify
wwlistand create bar chart that counts number of prospects purchased by race/ethnicity, separately for in-state vs. out-of-state
wwlist %>% filter(is.na(state)==0) %>% # drop obs where variable state missing
mutate( # create out-of-state indicator; create recoded ethnicity var
out_state = as_factor(if_else(state != "WA", "out-of-state", "in-state")),
ethn_race = recode(ethn_code,
"american indian or alaska native" = "nativeam",
"asian or native hawaiian or other pacific islander" = "api",
"black or african american" = "black",
"cuban" = "latinx",
"mexican/mexican american" = "latinx",
"not reported" = "not_reported",
"other-2 or more" = "multirace",
"other spanish/hispanic" = "latinx",
"puerto rican" = "latinx",
"white" = "white")) %>%
group_by(out_state) %>% # group_by "in-state" vs. "out-of-state"
count(ethn_race) %>% # count of number of prospects purchased by race
ggplot(aes(x=ethn_race, y=n)) + # plot
ylab("number of prospects") + xlab("race/ethnicity") +
geom_col() + coord_flip() + facet_wrap(~ out_state) Task:
- in one line of code, modify
wwlistand create bar chart of median income (in zip-code) of prospects purchased by race/ethnicity, separately for in-state vs. out-of-state
wwlist %>% filter(is.na(state)==0) %>% # drop obs where variable state missing
mutate( # create out-of-state indicator; create recoded ethnicity var
out_state = as_factor(if_else(state != "WA", "out-of-state", "in-state")),
ethn_race = recode(ethn_code,
"american indian or alaska native" = "nativeam",
"asian or native hawaiian or other pacific islander" = "api",
"black or african american" = "black",
"cuban" = "latinx",
"mexican/mexican american" = "latinx",
"not reported" = "not_reported",
"other-2 or more" = "multirace",
"other spanish/hispanic" = "latinx",
"puerto rican" = "latinx",
"white" = "white")) %>%
group_by(out_state, ethn_race) %>% # group_by "out-state" and ethnicity
summarize(avg_inc_zip = mean(med_inc_zip, na.rm = TRUE)) %>% # calculate avg. inc
ggplot(aes(x=out_state, y=avg_inc_zip)) +
ylab("avg. income in zip code") + xlab("") +
geom_col() + coord_flip() + facet_wrap(~ ethn_race) # plotExample R script from Ben Skinner, which creates analysis data for Skinner (2018)
Other relevant links
- Link to Github repository for Skinner (2018)
- Link to published paper
- Link to Skinner’s Github page
- A lot of cool stuff here
- Link to Skinner’s personal website
- A lot of cool stuff here
3.6 Which objects and functions are pipeable
Which objects and functions are “pipeable” (i.e., work with pipes)
- function is pipeable if it takes a data object as first argument and returns an object of same type
- In general, doesn’t seem to be any limit on which kinds of objects are pipeable (could be atomic vector, list, data frame)
# applying pipes to atomic vectors
1:10 %>% mean
#> [1] 5.5
1:10 %>% mean %>% str()
#> num 5.5But some pipeable functions restrict which kinds of data objects they accept
- In particular, the
dplyrfunctions (e.g.,filter,arrange, etc.) expect the first argument to be a data frame. dpylrfunctions won’t even accept a list as first argument, even though data frames are a particular class of list
wwlist %>% filter(firstgen == "Y") %>% str()
as.data.frame(wwlist) %>% str()
as.data.frame(wwlist) %>% filter(firstgen == "Y") %>% str()
as.list(wwlist) %>% str()
# as.list(wwlist) %>% filter(firstgen == "Y") %>% str() # error3.6.1 Practice exercise: Run the following code with and without pipes [Work through on your own]
Task:
- Count the number “first-generation” prospects from the state of Washington
Without pipes
count(filter(wwlist, firstgen == "Y", state == "WA"))
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 32428With pipes
wwlist %>% filter(firstgen == "Y", state == "WA") %>% count()
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 32428Task: frequency table of school_type for non first-gen prospects from WA
Without pipes
wwlist_temp <- filter(wwlist, firstgen == "N", state == "WA")
table(wwlist_temp$school_type, useNA = "always")
#>
#> private public <NA>
#> 11 46146 12489
rm(wwlist_temp) # cuz we don't need after creating tableWith pipes
wwlist %>% filter(firstgen == "N", state == "WA") %>% count(school_type)
#> # A tibble: 3 × 2
#> school_type n
#> <chr> <int>
#> 1 private 11
#> 2 public 46146
#> 3 <NA> 12489Comparison of two approaches
- without pipes, task requires multiple lines of code (this is quite common)
- first line creates object; second line analyzes object
- with pipes, task can be completed in one line of code and you aren’t left with objects you don’t care about
3.6.2 Student exercises with pipes
Using object
wwlistselect the following variables (state, firstgen, ethn_code) and assign<-them to objectwwlist_temp. (ex. wwlist_temp <- wwlist)Using the object you just created
wwlist_temp, create a frequency table ofethn_codefor first-gen prospects from California.Bonus: Try doing question 1 and 2 together. Use original object
wwlist, but do not assign to a new object.
Once finished you can rm(wwlist_temp)
Solutions pipes %>%
- Using object
wwlistselect the following variables (state, firstgen, ethn_code) and assign them to objectwwlist_temp
wwlist_temp <- wwlist %>%
select(state, firstgen, ethn_code) - Using the object you just created
wwlist_temp, create a frequency table ofethn_codefor first-gen prospects from California.
#names(wwlist)
wwlist_temp %>%
filter(firstgen == "Y", state == "CA") %>% count(ethn_code)
#> # A tibble: 10 × 2
#> ethn_code n
#> <chr> <int>
#> 1 american indian or alaska native 4
#> 2 asian or native hawaiian or other pacific islander 86
#> 3 black or african american 10
#> 4 cuban 1
#> 5 mexican/mexican american 643
#> 6 not reported 113
#> 7 other spanish/hispanic 179
#> 8 other-2 or more 4197
#> 9 puerto rican 8
#> 10 white 2933- Bonus: Try doing question 1 and 2 together.
wwlist %>%
select(state, firstgen, ethn_code) %>%
filter(firstgen == "Y", state == "CA") %>%
count(ethn_code)
#> # A tibble: 10 × 2
#> ethn_code n
#> <chr> <int>
#> 1 american indian or alaska native 4
#> 2 asian or native hawaiian or other pacific islander 86
#> 3 black or african american 10
#> 4 cuban 1
#> 5 mexican/mexican american 643
#> 6 not reported 113
#> 7 other spanish/hispanic 179
#> 8 other-2 or more 4197
#> 9 puerto rican 8
#> 10 white 2933
#rm(wwlist_temp)rm(wwlist_temp)4 Creating variables using mutate
We will load the data frame df_school_all. This data frame is based on data from the recruiting project. It contains one observation per US high school and variables identifying the number of visits by particular universities.
Data frame df_school_all
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_school_allvars.RData"))Our plan for learning how to create new variables
Recall that dplyr package within tidyverse provide a set of functions that can be described as “verbs”: subsetting, sorting, and transforming
| What we’ve done | Where we’re going |
|---|---|
| Subsetting data | Transforming data |
- select() variables |
- mutate() creates new variables |
- filter() observations |
- summarize() calculates across rows |
| Sorting data | - group_by() to calculate across rows within groups |
- arrange() |
Today
- We’ll use
mutate()to create new variables based on calculations across columns within a row
Next week
- We’ll combine
mutate()withsummarize()andgroup_by()to create variables based on calculations across rows
Create new data frame based on df_school_all
Recall, data frame df_school_all has one obs per US high school and then variables identifying number of visits by particular universities
names(df_school_all)
#> [1] "state_code" "school_type" "ncessch"
#> [4] "name" "address" "city"
#> [7] "zip_code" "pct_white" "pct_black"
#> [10] "pct_hispanic" "pct_asian" "pct_amerindian"
#> [13] "pct_other" "num_fr_lunch" "total_students"
#> [16] "num_took_math" "num_prof_math" "num_took_rla"
#> [19] "num_prof_rla" "avgmedian_inc_2564" "latitude"
#> [22] "longitude" "visits_by_196097" "visits_by_186380"
#> [25] "visits_by_215293" "visits_by_201885" "visits_by_181464"
#> [28] "visits_by_139959" "visits_by_218663" "visits_by_100751"
#> [31] "visits_by_199193" "visits_by_110635" "visits_by_110653"
#> [34] "visits_by_126614" "visits_by_155317" "visits_by_106397"
#> [37] "visits_by_149222" "visits_by_166629" "total_visits"
#> [40] "inst_196097" "inst_186380" "inst_215293"
#> [43] "inst_201885" "inst_181464" "inst_139959"
#> [46] "inst_218663" "inst_100751" "inst_199193"
#> [49] "inst_110635" "inst_110653" "inst_126614"
#> [52] "inst_155317" "inst_106397" "inst_149222"
#> [55] "inst_166629"Create new version of data frame, called school_v2, which we’ll use to introduce how to create new variables
school_v2 <- df_school_all %>%
select(-contains("inst_")) %>% # remove vars that start with "inst_"
rename( # rename selected variables
visits_by_berkeley = visits_by_110635,
visits_by_boulder = visits_by_126614,
visits_by_bama = visits_by_100751,
visits_by_stonybrook = visits_by_196097,
visits_by_rutgers = visits_by_186380,
visits_by_pitt = visits_by_215293,
visits_by_cinci = visits_by_201885,
visits_by_nebraska = visits_by_181464,
visits_by_georgia = visits_by_139959,
visits_by_scarolina = visits_by_218663,
visits_by_ncstate = visits_by_199193,
visits_by_irvine = visits_by_110653,
visits_by_kansas = visits_by_155317,
visits_by_arkansas = visits_by_106397,
visits_by_sillinois = visits_by_149222,
visits_by_umass = visits_by_166629,
num_took_read = num_took_rla,
num_prof_read = num_prof_rla,
med_inc = avgmedian_inc_2564
)
glimpse(school_v2)4.1 Introduce mutate() function
mutate() is dplyr approach to creating variables (not Base R approach)
Description of mutate()
- Creates new columns (variables) that are functions of existing columns
- After creating a new variable using
mutate(), every row of data is retained mutate()works best with pipes%>%
Investigate mutate() syntax
Usage (i.e., syntax)
mutate(.data,...)
Arguments
.data: a data frame- if using
mutate()after pipe operator%>%, then this argument can be omitted- Why? Because data frame object to left of
%>%“piped in” to first argument ofmutate()
- Why? Because data frame object to left of
- if using
...: expressions used to create new variables- “Name-value pairs of expressions”
- “The name of each argument will be the name of a new variable, and the value will be its corresponding value.”
- “Use a
NULLvalue in mutate to drop a variable.” - “New variables overwrite existing variables of the same name”
Value
- returns a (data frame) object that contains the original input data frame and new variables that were created by
mutate()
Task:
- Using data frame
school_v2create new variable that measures the pct of students on free/reduced lunch (output omitted)
# create new dataset with fewer vars; not necessary to do this
school_sml <- school_v2 %>%
select(ncessch, school_type, num_fr_lunch, total_students)
#print obs
head(school_sml, 10)
# create new var
school_sml %>%
mutate(pct_fr_lunch = num_fr_lunch/total_students)
#362/501
# remove data frame object
rm(school_sml) Can create variables using standard mathematical or logical operators [output omitted]
#glimpse(school_v2)
school_v2 %>%
select(state_code,school_type,ncessch,med_inc,num_fr_lunch,total_students,num_took_math) %>%
mutate( # each argument creates a new variable, name of argument is name of variable
one = 1,
med_inc000 = med_inc/1000,
pct_fr_lunch = num_fr_lunch/total_students*100,
took_math_na = is.na(num_took_math)==1
) %>%
select(state_code,school_type,ncessch,one,med_inc,med_inc000,num_fr_lunch,total_students,pct_fr_lunch,num_took_math,took_math_na)Can create variables using “helper functions” called within mutate() [output omitted]
- These are standalone functions can be called within
mutate()- e.g.,
if_else(),recode(),case_when()
- e.g.,
- will walk through helper functions in more detail in subsequent sections of lecture
school_v2 %>%
select(state_code,ncessch,name,school_type) %>%
mutate(public = if_else(school_type == "public", 1, 0))New variable not retained unless we assign <- it to an object (existing or new)
mutate()without assignment
school_v2 %>% mutate(pct_fr_lunch = num_fr_lunch/total_students)
names(school_v2)mutate()with assignment
school_v2_temp <- school_v2 %>%
mutate(pct_fr_lunch = num_fr_lunch/total_students)
names(school_v2_temp)
rm(school_v2_temp)4.2 mutate() to create multiple variables at once
school_v2 %>%
mutate(pct_fr_lunch = num_fr_lunch/total_students,
pct_prof_math= num_prof_math/num_took_math) %>%
select(num_fr_lunch, total_students, pct_fr_lunch,
num_prof_math, num_took_math, pct_prof_math)Or we could write code this way:
school_v2 %>%
select(num_fr_lunch, total_students, num_prof_math, num_took_math) %>%
mutate(pct_fr_lunch = num_fr_lunch/total_students,
pct_prof_math= num_prof_math/num_took_math) mutate() can use variables previously created within mutate()
school_v2 %>%
select(num_prof_math, num_took_math, num_took_read,num_prof_read) %>%
mutate(pct_prof_math = num_prof_math/num_took_math,
pct_prof_read = num_prof_read/num_took_read,
avg_pct_prof_math_read = (pct_prof_math + pct_prof_read)/2) 4.3 mutate(), removing variables created by mutate()
Within mutate() use syntax var_name = NULL to remove variable from data frame
- note: Variable not permanently removed from data frame unless you use assignment
<-to create new data frame or overwrite existing data frame
ncol(school_v2)
school_v2 %>%
select(num_prof_math, num_took_math, num_took_read,num_prof_read) %>% glimpse()
school_v2 %>%
select(num_prof_math, num_took_math, num_took_read,num_prof_read) %>%
mutate(num_prof_math = NULL, num_took_math = NULL) %>% glimpse()
#But variables not permanently removed because we didn't use assignment
ncol(school_v2)Why would we remove variables within mutate() rather select()?
- remove temporary “work” variables used to create desired variable
- Example: measure of average of pct who passed math and pct who passed reading
school_v2 %>%
select(num_prof_math, num_took_math, num_took_read,num_prof_read) %>%
mutate(pct_prof_math = num_prof_math/num_took_math, # create work var
pct_prof_read = num_prof_read/num_took_read, # create work var
avg_pct_prof_math_read = (pct_prof_math + pct_prof_read)/2, #create analysis var
pct_prof_math = NULL, # remove work var
pct_prof_read = NULL) %>% # remove work var
glimpse()4.3.1 Student exercise using mutate()
Using the object
school_v2, select the following variables (num_prof_math,num_took_math,num_prof_read,num_took_read) and create a measure of percent proficient in mathpct_prof_mathand percent proficient in readingpct_prof_read.Now using the code for question 1, filter schools where at least 50% of students are proficient in math & reading.
Count the number of schools from question 2.
Using
school_v2, usingmutate()combined withis.na()create a dichotomous indicator variablemed_inc_nathat identifies whethermed_incis missing (NA) or not. And then use syntaxcount(var_name)to create frequency table of variablemed_inc_na. How many observations are missing?
Solutions for exercise using mutate()
- Using the object
school_v2, select the following variables (num_prof_math,num_took_math,num_prof_read,num_took_read) and create a measure of percent proficient in mathpct_prof_mathand percent proficient in readingpct_prof_read.
school_v2 %>%
select(num_prof_math, num_took_math, num_prof_read, num_took_read) %>%
mutate(pct_prof_math = num_prof_math/num_took_math,
pct_prof_read = num_prof_read/num_took_read)
#> # A tibble: 21,301 × 6
#> num_prof_math num_took_math num_prof_read num_took_read pct_prof_math
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 24.8 146 25.0 147 0.17
#> 2 1.7 17 1.7 17 0.1
#> 3 3.5 14 3.5 14 0.25
#> 4 3 30 3 30 0.1
#> 5 2.8 28 2.8 28 0.1
#> 6 2.5 25 2.4 24 0.1
#> 7 1.55 62 1.55 62 0.025
#> 8 2.1 21 2.2 22 0.1
#> 9 2.3 23 2.3 23 0.1
#> 10 1.9 19 1.9 19 0.1
#> # ℹ 21,291 more rows
#> # ℹ 1 more variable: pct_prof_read <dbl>- Now using the code for question 1, filter schools where at least 50% of students are proficient in math & reading.
school_v2 %>%
select(num_prof_math, num_took_math, num_prof_read, num_took_read) %>%
mutate(pct_prof_math = num_prof_math/num_took_math,
pct_prof_read = num_prof_read/num_took_read) %>%
filter(pct_prof_math >= 0.5 & pct_prof_read >= 0.5)
#> # A tibble: 7,760 × 6
#> num_prof_math num_took_math num_prof_read num_took_read pct_prof_math
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 135. 260 149. 261 0.52
#> 2 299. 475 418 475 0.63
#> 3 213. 410 332. 410 0.52
#> 4 54.6 105 96.6 105 0.52
#> 5 111. 121 118. 121 0.92
#> 6 1057. 1994 1477. 2204 0.53
#> 7 100. 103 125. 128 0.975
#> 8 56.4 99 84.4 148 0.57
#> 9 445. 586 392. 594 0.76
#> 10 56.0 59 53.1 61 0.95
#> # ℹ 7,750 more rows
#> # ℹ 1 more variable: pct_prof_read <dbl>- Count the number of schools from question 2.
school_v2 %>%
select(num_prof_math, num_took_math, num_prof_read, num_took_read) %>%
mutate(pct_prof_math = num_prof_math/num_took_math,
pct_prof_read = num_prof_read/num_took_read) %>%
filter(pct_prof_math >= 0.5 & pct_prof_read >= 0.5) %>%
count()
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 7760- Using
school_v2, usingmutate()combined withis.na()create a dichotomous indicator variablemed_inc_nathat identifies whethermed_incis missing (NA) or not. And then use syntaxcount(var_name)to create frequency table of variablemed_inc_na. How many observations are missing?
school_v2 %>%
mutate(med_inc_na = is.na(med_inc)) %>%
count(med_inc_na)
#> # A tibble: 2 × 2
#> med_inc_na n
#> <lgl> <int>
#> 1 FALSE 20677
#> 2 TRUE 6244.4 Using if_else() function within mutate()
Description
- if
<condition>TRUE, assign value; if<condition>FALSEassign value
Usage (i.e., syntax)
if_else(logical condition, true, false, missing = NULL)
Arguments
logical condition: a condition that evaluates toTRUEorFALSEtrue: value to assign if conditionTRUEfalse: value to assign if conditionFALSEmissing: value to assign to rows that have valueNAfor condition- default is
missing = NULL; means that if condition isNA, then new_var ==NA - But can assign different values to
NAs, e.g.,missing = -9
- default is
Value
- “Where condition is TRUE, the matching value from true, where it’s FALSE, the matching value from false, otherwise NA.”
- Unless otherwise specified,
NAs in “input” var(s) assignedNAin “output var”
Example: Create 0/1 indicator of whether got at least one visit from Berkeley
school_v2 %>%
mutate(got_visit_berkeley = if_else(visits_by_berkeley>0,1,0)) %>%
count(got_visit_berkeley)4.4.1 if_else() within mutate() to create 0/1 indicator variables
We often create dichotomous (0/1) indicator variables of whether something happened (or whether something is TRUE)
- Variables that are of substantive interest to project
- e.g., did student graduate from college
- Variables that help you investigate data, check quality
- e.g., indicator of whether an observation is missing/non-missing for a particular variable
Task
- Create 0/1 indicator if school has median income greater than $100,000
Usually a good idea to investigate “input” variables before creating analysis vars
str(school_v2$med_inc) # investigate variable type
school_v2 %>% count(med_inc) # frequency count, but this isn't very helpful
school_v2 %>% filter(is.na(med_inc)) %>% count()
# shows number of obs w/ missing med_incCreate variable
school_v2 %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0)) %>%
count(inc_gt_100k) # note how NA values of med_inc treated
#> # A tibble: 3 × 2
#> inc_gt_100k n
#> <dbl> <int>
#> 1 0 18632
#> 2 1 2045
#> 3 NA 624Task:
- Create 0/1 indicator if school has median income greater than $100,000.
This time, let’s experiment with the missing argument of if_else()
#what we wrote before
school_v2 %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0)) %>%
count(inc_gt_100k)
#manually write out the default value for `missing`
school_v2 %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0, missing = NULL)) %>%
count(inc_gt_100k) # note how NA values of med_inc treated
school_v2 %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0, missing = NA_real_)) %>%
count(inc_gt_100k) # note how NA values of med_inc treated
# NA can be coerced to any other vector type except raw:
# NA_integer_, NA_real_, NA_complex_ and NA_character_
# Here we give missing values in condition the value of -9 in new variable
school_v2 %>% select(med_inc) %>%
mutate(inc_gt_100k= if_else(med_inc>100000,1,0, missing = -9)) %>%
count(inc_gt_100k) Task
- Create 0/1 indicator variable
nonmiss_mathwhich indicates whether school has non-missing values for the variablenum_took_math- note:
num_took_mathrefers to number of students at a school that took state math proficiency test
- note:
Usually a good to investigate “input” variables before creating analysis vars
school_v2 %>% count(num_took_math) # this isn't very helpful
school_v2 %>% filter(is.na(num_took_math)) %>% count(num_took_math) # shows number of obs w/ missing num_took_mathCreate variable
school_v2 %>% select(num_took_math) %>%
mutate(nonmiss_math= if_else(!is.na(num_took_math),1,0)) %>%
count(nonmiss_math) # note how NA values treated
#> # A tibble: 2 × 2
#> nonmiss_math n
#> <dbl> <int>
#> 1 0 4103
#> 2 1 171984.4.2 Student exercises if_else()
- Using the object
school_v2, create 0/1 indicator variablein_state_berkeleythat equals1if the high school is in the same state as UC Berkeley (i.e.,state_code=="CA").
- Create 0/1 indicator
berkeley_and_irvineof whether a school got at least one visit from UC Berkeley AND from UC Irvine.
- Create 0/1 indicator
berkeley_or_irvineof whether a school got at least one visit from UC Berkeley OR from UC Irvine.
Solutions if_else()
- Using the object
school_v2, create 0/1 indicator variablein_state_berkeleythat equals1if the high school is in the same state as UC Berkeley (i.e.,state_code=="CA").
str(school_v2$state_code) # investigate input variable
school_v2 %>% filter(is.na(state_code)) %>% count() # investigate input var
#Create var
school_v2 %>% mutate(in_state_berkeley=if_else(state_code=="CA",1,0)) %>%
count(in_state_berkeley)- Create 0/1 indicator
berkeley_and_irvineof whether a school got at least one visit from UC Berkeley AND from UC Irvine.
#investigate input vars
school_v2 %>% select(visits_by_berkeley, visits_by_irvine) %>% str()
school_v2 %>% filter(is.na(visits_by_berkeley)) %>% count()
school_v2 %>% filter(is.na(visits_by_irvine)) %>% count()
#create variable
school_v2 %>%
mutate(berkeley_and_irvine=if_else(visits_by_berkeley>0
& visits_by_irvine>0,1,0)) %>%
count(berkeley_and_irvine)- Create 0/1 indicator
berkeley_or_irvineof whether a school got at least one visit from UC Berkeley OR from UC Irvine.
school_v2 %>%
mutate(berkeley_or_irvine=if_else(visits_by_berkeley>0 | visits_by_irvine>0,1,0)) %>%
count(berkeley_or_irvine)4.5 Using recode() function within mutate()
Description: Recodes values of a variable
Usage (i.e., syntax)
- recode(.x, …, .default = NULL, .missing = NULL)
Arguments [see help file for further details]
.xA vector (e.g., variable) to modify...Specifications for recode, of formcurrent_value = new_recoded_value.default: If supplied, all values not otherwise matched given this value..missing: If supplied, any missing values in .x replaced by this value.
Example: Using data frame wwlist, create new 0/1 indicator public_school from variable school_type
str(wwlist$school_type)
wwlist %>% count(school_type)
wwlist_temp <- wwlist %>% select(school_type) %>%
mutate(public_school = recode(school_type,"public" = 1, "private" = 0))
wwlist_temp %>% head(n=10)
str(wwlist_temp$public_school) # note: numeric variable
wwlist_temp %>% count(public_school) # note the NAs
rm(wwlist_temp)Recoding school_type could have been accomplished using if_else()
- Use
recode()when new variable has more than two categories
Task: Create school_catv2 based on school_category with these categories:
- “regular”; “alternative”; “special”; “vocational”
Investigate input var
str(wwlist$school_category) # character variable
wwlist %>% count(school_category) Recode
wwlist_temp <- wwlist %>% select(school_category) %>%
mutate(school_catv2 = recode(school_category,
"Alternative Education School" = "alternative",
"Alternative/other" = "alternative",
"Regular elementary or secondary" = "regular",
"Regular School" = "regular",
"Special Education School" = "special",
"Special program emphasis" = "special",
"Vocational Education School" = "vocational")
)
str(wwlist_temp$school_catv2) # character variable created
wwlist_temp %>% count(school_catv2)
rm(wwlist_temp)Task: Create school_catv2 based on school_category with these categories:
- “regular”; “alternative”; “special”; “vocational”
- This time use the
.missingargument to recodeNAsto “unknown”
wwlist_temp <- wwlist %>% select(school_category) %>%
mutate(school_catv2 = recode(school_category,
"Alternative Education School" = "alternative",
"Alternative/other" = "alternative",
"Regular elementary or secondary" = "regular",
"Regular School" = "regular",
"Special Education School" = "special",
"Special program emphasis" = "special",
"Vocational Education School" = "vocational",
.missing = "unknown")
)
str(wwlist_temp$school_catv2)
wwlist_temp %>% count(school_catv2)
wwlist %>% count(school_category)
rm(wwlist_temp)Task: Create school_catv2 based on school_category with these categories:
- “regular”; “alternative”; “special”; “vocational”
- This time use the
.defaultargument to assign the value “regular”
wwlist_temp <- wwlist %>% select(school_category) %>%
mutate(school_catv2 = recode(school_category,
"Alternative Education School" = "alternative",
"Alternative/other" = "alternative",
"Special Education School" = "special",
"Special program emphasis" = "special",
"Vocational Education School" = "vocational",
.default = "regular")
)
str(wwlist_temp$school_catv2)
wwlist_temp %>% count(school_catv2)
wwlist %>% count(school_category)
rm(wwlist_temp)Task: Create school_catv2 based on school_category with these categories:
- This time create a numeric variable rather than character:
1for “regular”;2for “alternative”;3for “special”;4for “vocational”
wwlist_temp <- wwlist %>% select(school_category) %>%
mutate(school_catv2 = recode(school_category,
"Alternative Education School" = 2,
"Alternative/other" = 2,
"Regular elementary or secondary" = 1,
"Regular School" = 1,
"Special Education School" = 3,
"Special program emphasis" = 3,
"Vocational Education School" = 4)
)
str(wwlist_temp$school_catv2) # note: numeric variable now
wwlist_temp %>% count(school_catv2)
wwlist %>% count(school_category)
rm(wwlist_temp)4.5.1 Student exercise using recode() within mutate()
load(url("https://github.com/ozanj/rclass/raw/master/data/recruiting/recruit_event_somevars.RData"))
names(df_event)- Using object
df_event, assign new objectdf_event_tempand a numeric variable createevent_typev2based onevent_typewith these categories:1for “2yr college”;2for “4yr college”;3for “other”;4for “private hs”;5for “public hs”
- This time use the
.defaultargument to assign the value5for “public hs”
Solutions using recode() within mutate()
Check input variable
names(df_event)
str(df_event$event_type)
df_event %>% count(event_type)- Using object
df_event, assign new objectdf_event_tempand create a numeric variableevent_typev2based onevent_typewith these categories:1for “2yr college”;2for “4yr college”;3for “other”;4for “private hs”;5for “public hs”
df_event_temp <- df_event %>%
select(event_type) %>%
mutate(event_typev2 = recode(event_type,
"2yr college" = 1,
"4yr college" = 2,
"other" = 3,
"private hs" = 4,
"public hs" = 5)
)
str(df_event_temp$event_typev2)
df_event_temp %>% count(event_typev2)
df_event %>% count(event_type)- This time assign the value use the
.defaultargument to assign the value5for “public hs”
df_event_temp <- df_event %>% select(event_type) %>%
mutate(event_typev2 = recode(event_type,
"2yr college" = 1,
"4yr college" = 2,
"other" = 3,
"private hs" = 4,
.default = 5)
)
str(df_event_temp$event_typev2)
df_event_temp %>% count(event_typev2)
df_event %>% count(event_type)4.6 Using case_when() function within mutate()
case_when() useful for creating variable that is a function of multiple “input” variables
Usage (i.e., syntax): case_when(...)
Arguments [from help file; see help file for more details]
...: A sequence of two-sided formulas.- The left hand side (LHS) determines which values match this case.
- LHS must evaluate to a logical vector.
- LHS must evaluate to a logical vector.
- The right hand side (RHS) provides the replacement value.
- The left hand side (LHS) determines which values match this case.
Task: Using data frame wwlist and input vars state and firstgen, create a 4-category var with following categories:
- “instate_firstgen”; “instate_nonfirstgen”; “outstate_firstgen”; “outstate_nonfirstgen”
wwlist_temp <- wwlist %>% select(state,firstgen) %>%
mutate(state_gen = case_when(
state == "WA" & firstgen =="Y" ~ "instate_firstgen",
state == "WA" & firstgen =="N" ~ "instate_nonfirstgen",
state != "WA" & firstgen =="Y" ~ "outstate_firstgen",
state != "WA" & firstgen =="N" ~ "outstate_nonfirstgen")
)
str(wwlist_temp$state_gen)
wwlist_temp %>% count(state_gen)Let’s take a closer look at how values of inputs are coded into values of outputs
wwlist %>% select(state,firstgen) %>% str()
count(wwlist,state)
count(wwlist,firstgen)Create variable
wwlist_temp <- wwlist %>% select(state,firstgen) %>%
mutate(state_gen = case_when(
state == "WA" & firstgen =="Y" ~ "instate_firstgen",
state == "WA" & firstgen =="N" ~ "instate_nonfirstgen",
state != "WA" & firstgen =="Y" ~ "outstate_firstgen",
state != "WA" & firstgen =="N" ~ "outstate_nonfirstgen")
)Compare values of input vars to value of output var
wwlist_temp %>% count(state_gen)
wwlist_temp %>% filter(is.na(state)) %>% count(state_gen)
wwlist_temp %>% filter(is.na(firstgen)) %>% count(state_gen)
wwlist_temp %>% filter(is.na(firstgen) | is.na(state)) %>% count(state_gen)Take-away: by default var created by case_when() equals NA for obs where one of the inputs equals NA
4.6.1 Student exercise using case_when() within mutate()
- Using the object
school_v2and input varsschool_type, andstate_code, create a 4-category varstate_typewith following categories:- “instate_public”; “instate_private”; “outstate_public”; “outstate_private”
- Note: We are referring to CA as in-state for this example
Solutions using case_when() within mutate()
Investigate
school_v2 %>% select(state_code,school_type) %>% str()
count(school_v2,state_code)
school_v2 %>% filter(is.na(state_code)) %>% count()
count(school_v2,school_type)
school_v2 %>% filter(is.na(school_type)) %>% count()Using the object
school_v2and input varsschool_type, andstate_code, create a 4-category varstate_typewith following categories:- “instate_public”; “instate_private”; “outstate_public”; “outstate_private”
school_v2_temp <- school_v2 %>% select(state_code,school_type) %>%
mutate(state_type = case_when(
state_code == "CA" & school_type == "public" ~ "instate_public",
state_code == "CA" & school_type == "private" ~ "instate_private",
state_code != "CA" & school_type == "public" ~ "outstate_public",
state_code != "CA" & school_type == "private" ~ "outstate_private")
)
school_v2_temp %>% count(state_type)
#> # A tibble: 4 × 2
#> state_type n
#> <chr> <int>
#> 1 instate_private 366
#> 2 instate_public 1404
#> 3 outstate_private 3456
#> 4 outstate_public 16075
#school_v2_temp %>% filter(is.na(state_code)) %>% count(state_type) #no missing
#school_v2_temp %>% filter(is.na(school_type)) %>% count(state_type) #no missing