library(tidyverse)
library(haven)
library(labelled)
library(lubridate)Attributes and Class
1 Introduction
1.1 Libraries we will use
Load the packages we will use by running this code chunk:
If package not yet installed, then must install before you load. Install in “console” rather than .Rmd file:
- Generic syntax:
install.packages("package_name") - Install “tidyverse”:
install.packages("tidyverse")
Note: When we load package, name of package is not in quotes; but when we install package, name of package is in quotes:
install.packages("tidyverse")library(tidyverse)
1.2 Dataset we will use
Recall from Into the tidyverse lecture, we used the data set wwlist. This data set represents “prospects” purchased by Western Washington U.
- Essentially, universities identify/target “prospect” by buying student lists from College Board/ACT (e.g., $.50+ per prospect).
- Data universities purchase on students includes their contact info (e.g., address, email), academics (e.g., GPA, AP coursework), socioeconomic and demographic characteristics.
- Universities choose which prospective students’ information to purchase by filtering on criteria like their zip code, GPA, test score range, etc.
rm(list = ls()) # remove all objects
load(url("https://github.com/anyone-can-cook/rclass1/raw/master/data/prospect_list/wwlist_merged.RData"))2 Attributes and augmented vectors
2.1 Review data structures: Vectors
Two types of vectors:
- Atomic vectors
- Lists
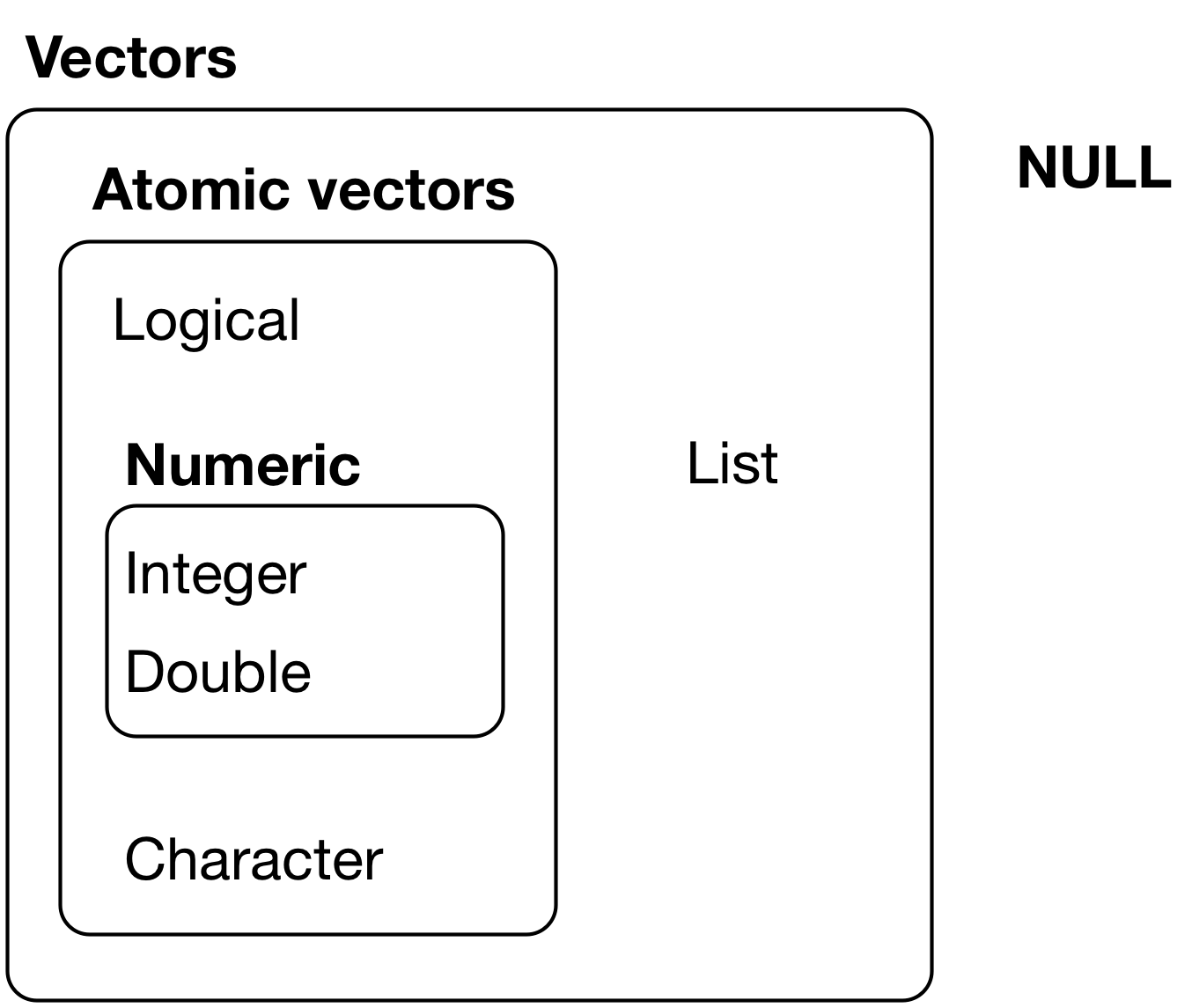
2.1.1 Atomic vectors
An atomic vector is a collection of values
- Each value in an atomic vector is an element
- All elements within vector must have the same data type
(a <- c(1,2,3)) # parentheses () assign and print object in one step[1] 1 2 3length(a) # length = number of elements[1] 3typeof(a) # numeric atomic vector, type=double[1] "double"str(a) # investigate structure of object num [1:3] 1 2 3Can assign names to vector elements, creating a named atomic vector
(b <- c(v1=1,v2=2,v3=3))v1 v2 v3
1 2 3 length(b) [1] 3typeof(b) [1] "double"str(b) Named num [1:3] 1 2 3
- attr(*, "names")= chr [1:3] "v1" "v2" "v3"2.1.2 Lists
- Like atomic vectors, lists are objects that contain elements
- However, data type can differ across elements within a list
- E.g., an element of a list can be another list
list_a <- list(1,2,"apple")
typeof(list_a)[1] "list"length(list_a)[1] 3str(list_a)List of 3
$ : num 1
$ : num 2
$ : chr "apple"list_b <- list(1, c("apple", "orange"), list(1, 2))
length(list_b)[1] 3str(list_b)List of 3
$ : num 1
$ : chr [1:2] "apple" "orange"
$ :List of 2
..$ : num 1
..$ : num 2Like atomic vectors, elements within a list can be named, thereby creating a named list
# not named
str(list_b) List of 3
$ : num 1
$ : chr [1:2] "apple" "orange"
$ :List of 2
..$ : num 1
..$ : num 2# named
list_c <- list(v1=1, v2=c("apple", "orange"), v3=list(1, 2, 3))
str(list_c) List of 3
$ v1: num 1
$ v2: chr [1:2] "apple" "orange"
$ v3:List of 3
..$ : num 1
..$ : num 2
..$ : num 32.1.3 Data frames
A data frame is a list with the following characteristics:
- All the elements must be vectors with the same length
- Data frames are augmented lists because they have additional attributes
# a regular list
(list_d <- list(col_a = c(1,2,3), col_b = c(4,5,6), col_c = c(7,8,9)))$col_a
[1] 1 2 3
$col_b
[1] 4 5 6
$col_c
[1] 7 8 9typeof(list_d)[1] "list"attributes(list_d)$names
[1] "col_a" "col_b" "col_c"# a data frame
(df_a <- data.frame(col_a = c(1,2,3), col_b = c(4,5,6), col_c = c(7,8,9))) col_a col_b col_c
1 1 4 7
2 2 5 8
3 3 6 9typeof(df_a)[1] "list"attributes(df_a)$names
[1] "col_a" "col_b" "col_c"
$class
[1] "data.frame"
$row.names
[1] 1 2 32.2 Converting between types
Functions for converting between types:
as.logical(): Convert tologicalas.numeric(): Convert tonumericas.integer(): Convert tointegeras.character(): Convert tocharacteras.list(): Convert tolistas.data.frame(): Convert todata.frame
Before converting between vectors types, it is important to know that the most complex type always wins.
typeof(c(TRUE, 3L))[1] "integer"#> [1] "integer"
typeof(c(2L, 4.5))[1] "double"#> [1] "double"
typeof(c(2.5, "j"))[1] "character"#> [1] "character"Credit: R for Data Science, section on Coercion
Convert to logical using as.logical()
Character vector coerced to logical vector:
# Only "TRUE"/"FALSE", "True"/"False", "T"/"F", "true"/"false" are able to be coerced to logical type
as.logical(c("TRUE", "FALSE", "True", "False", "true", "false", "T", "F", "t", "f", "")) [1] TRUE FALSE TRUE FALSE TRUE FALSE TRUE FALSE NA NA NANumeric vector coerced to logical vector:
# 0 is treated as FALSE, while all other numeric values are treated as TRUE
as.logical(c(0, 0.0, 1, -1, 20, 5.5))[1] FALSE FALSE TRUE TRUE TRUE TRUEConvert to numeric using as.numeric()
Logical vector coerced to numeric vector:
# FALSE is mapped to 0 and TRUE is mapped to 1
as.numeric(c(FALSE, TRUE))[1] 0 1Character vector coerced to numeric vector:
# Strings containing numeric values can be coerced to numeric (leading 0's are dropped)
# All other characters become NA
as.numeric(c("0", "007", "2.5", "abc", "."))[1] 0.0 7.0 2.5 NA NAConvert to integer using as.integer()
Logical vector coerced to integer vector:
# FALSE is mapped to 0 and TRUE is mapped to 1
as.integer(c(FALSE, TRUE))[1] 0 1Character vector coerced to integer vector:
# Strings containing numeric values can be coerced to integer (leading 0's are dropped, decimals are truncated)
# All other characters become NA
as.integer(c("0", "007", "2.5", "abc", "."))[1] 0 7 2 NA NANumeric vector coerced to integer vector:
# All decimal places are truncated
as.integer(c(0, 2.1, 10.5, 8.8, -1.8))[1] 0 2 10 8 -1Convert to character using as.character()
Logical vector coerced to character vector:
as.character(c(FALSE, TRUE))[1] "FALSE" "TRUE" Numeric vector coerced to character vector:
as.character(c(-5, 0, 2.5))[1] "-5" "0" "2.5"Integer vector coerced to character vector:
as.character(c(-2L, 0L, 10L))[1] "-2" "0" "10"Convert to list using as.list()
Atomic vectors coerced to list:
# Logical vector
as.list(c(TRUE, FALSE))[[1]]
[1] TRUE
[[2]]
[1] FALSE# Character vector
as.list(c("a", "b", "c"))[[1]]
[1] "a"
[[2]]
[1] "b"
[[3]]
[1] "c"# Numeric vector
as.list(1:3)[[1]]
[1] 1
[[2]]
[1] 2
[[3]]
[1] 3Convert to data.frame using as.data.frame()
Lists coerced to dataframe:
# Create a list
l <- list(A = c("x", "y", "z"), B = c(1, 2, 3))
str(l)List of 2
$ A: chr [1:3] "x" "y" "z"
$ B: num [1:3] 1 2 3# Convert to class `data.frame`
df <- as.data.frame(l, stringsAsFactors = F)
str(df)'data.frame': 3 obs. of 2 variables:
$ A: chr "x" "y" "z"
$ B: num 1 2 3Example: Practical example of converting type
When working with data, it may be helpful to label values for certain variables. Data files often come with a code book that defines how values are coded. Let’s look at an example of labeling values and how converting data type may come into play.
We’ll look at the FIPS variable from the Integrated Postsecondary Education Data System (IPEDS) data. The state FIPS code is a numeric code that identifies a state. For example, 1 is the FIPS code for Alabama, 2 is the FIPS code for Alaska, etc. We’ll want to label each numeric value in the FIPS column with the corresponding state name.
# Library for labeling variables and values in a dataframe
library(labelled)
# Read in IPEDS data and codebook
ipeds_df <- read.csv('https://raw.githubusercontent.com/cyouh95/recruiting-chapter/master/data/ipeds_hd2017.csv', header = TRUE, na.strings=c('', 'NA'), stringsAsFactors = F)
ipeds_values <- read.csv('https://raw.githubusercontent.com/cyouh95/recruiting-chapter/master/data/ipeds_hd2017_values.csv', header = TRUE, na.strings=c('', 'NA'), stringsAsFactors = F)
# The codebook defines how variables are coded, such as STABBR, FIPS, and other variables
head(ipeds_values) varnumber varname codevalue valuelabel frequency percent
1 10016 STABBR AL Alabama 95 1.33
2 10016 STABBR AK Alaska 10 0.14
3 10016 STABBR AZ Arizona 137 1.92
4 10016 STABBR AR Arkansas 86 1.20
5 10016 STABBR CA California 729 10.19
6 10016 STABBR CO Colorado 118 1.65# Filter codebook for just the values for the FIPS variable
fips_values <- ipeds_values %>% filter(varname == 'FIPS') %>% select(varname, codevalue, valuelabel)
head(fips_values) varname codevalue valuelabel
1 FIPS 1 Alabama
2 FIPS 2 Alaska
3 FIPS 4 Arizona
4 FIPS 5 Arkansas
5 FIPS 6 California
6 FIPS 8 Colorado
When we read in the data from the CSV files, R automatically tries to determine the data type of each variable. As seen below, the FIPS column from the ipeds_df that we want to label is of type integer, while the codevalue column from the codebook is of type character (since not all values are numeric):
# Type of `FIPS` column
str(ipeds_df$FIPS) int [1:7153] 1 1 1 1 1 1 1 1 1 1 ...# Type of `codevalue` column
str(fips_values$codevalue) chr [1:59] "1" "2" "4" "5" "6" "8" "9" "10" "11" "12" "13" "15" "16" "17" ...
This discrepancy becomes a problem when we try to label the value using the labelled library:
# Error: `x` and `labels` must be same type
val_label(ipeds_df$FIPS, fips_values[1, 'codevalue']) <- fips_values[1, 'valuelabel']
To resolve this, we can use as.integer() to convert the codevalue from character type to integer before trying to label the value:
# This now works
val_label(ipeds_df$FIPS, as.integer(fips_values[1, 'codevalue'])) <- fips_values[1, 'valuelabel']
# Check value labels
val_labels(ipeds_df$FIPS)Alabama
1 # We can use as.integer() to convert the entire vector (ie. codevalue column) to integer
fips_values$codevalue <- as.integer(fips_values$codevalue)
# Type of `codevalue` column
str(fips_values$codevalue) int [1:59] 1 2 4 5 6 8 9 10 11 12 ...# Use loop to label the rest of the values
for (i in 1:nrow(fips_values)) {
val_label(ipeds_df$FIPS, fips_values[i, 'codevalue']) <- fips_values[i, 'valuelabel']
}
# Check value labels
val_labels(ipeds_df$FIPS) Alabama Alaska
1 2
Arizona Arkansas
4 5
California Colorado
6 8
Connecticut Delaware
9 10
District of Columbia Florida
11 12
Georgia Hawaii
13 15
Idaho Illinois
16 17
Indiana Iowa
18 19
Kansas Kentucky
20 21
Louisiana Maine
22 23
Maryland Massachusetts
24 25
Michigan Minnesota
26 27
Mississippi Missouri
28 29
Montana Nebraska
30 31
Nevada New Hampshire
32 33
New Jersey New Mexico
34 35
New York North Carolina
36 37
North Dakota Ohio
38 39
Oklahoma Oregon
40 41
Pennsylvania Rhode Island
42 44
South Carolina South Dakota
45 46
Tennessee Texas
47 48
Utah Vermont
49 50
Virginia Washington
51 53
West Virginia Wisconsin
54 55
Wyoming American Samoa
56 60
Federated States of Micronesia Guam
64 66
Marshall Islands Northern Marianas
68 69
Palau Puerto Rico
70 72
Virgin Islands
78 2.3 Atomic vectors versus augmented vectors
Atomic vectors [our focus so far]
- I think of atomic vectors as “just the data”
- Atomic vectors are the building blocks for augmented vectors
Augmented vectors
- Augmented vectors are atomic vectors with additional attributes attached
Attributes
- Attributes are additional “metadata” that can be attached to any object (e.g., vector or list)
Example: Variables of a dataset
- A data frame is a list
- Each element in the list is a variable, which consists of:
- Atomic vector (“just the data”)
- Any attributes we want to attach to each element/variable
- Variable name, an attribute of the data frame object
Other examples of attributes in R
- Value labels: Character labels (e.g., “Charter School”) attached to numeric values
- Object class: Specifies how object is treated by object oriented programming language
Main takeaway:
- Augmented vectors are atomic vectors (just the data) with additional attributes attached
2.4 Attributes and functions to identify/modify attributes
Description of attributes from Grolemund and Wickham 20.6
- “Any vector can contain arbitrary additional metadata through its attributes”
- “You can think of attributes as named list of vectors that can be attached to any object”
Functions to identify and modify attributes
attributes()function to describe all attributes of an objectattr()to see individual attribute of an object or set/change an individual attribute of an object
2.4.1 Describe all attributes of an object
# pull up help file for the attributes() function
?attributesAttributes of a named atomic vector:
# create named atomic vector
(vector1 <- c(a = 1, b = 2, c = 3, d = 4))a b c d
1 2 3 4 attributes(vector1)$names
[1] "a" "b" "c" "d"attributes(vector1) %>% str() # a named list of vectors!List of 1
$ names: chr [1:4] "a" "b" "c" "d"# remove all attributes from the object
attributes(vector1) <- NULL
vector1[1] 1 2 3 4attributes(vector1)NULL2.4.2 Attributes of a variable in a data frame
Accessing variable using [[]] subset operator
- Recall
object_name[["element_name"]]accesses contents of the element - If object is a data frame,
df_name[["var_name"]]accesses contents of variable- For simple vars like
firstgen, syntax yields an atomic vector (“just the data”)
- For simple vars like
- Shorthand syntax for
df_name[["var_name"]]isdf_name$var_name
str(wwlist[["firstgen"]]) chr [1:268396] NA "N" "N" "N" NA "N" "N" "Y" "Y" "N" "N" "N" "N" "N" "N" ...attributes(wwlist[["firstgen"]])NULLstr(wwlist$firstgen) # same same chr [1:268396] NA "N" "N" "N" NA "N" "N" "Y" "Y" "N" "N" "N" "N" "N" "N" ...attributes(wwlist$firstgen)NULLAccessing variable using [] subset operator
object_name["element_name"]creates object of same type asobject_name- If object is a data frame,
df_name["var_name"]returns a data frame containing just thevar_namecolumn
str(wwlist["firstgen"])
attributes(wwlist["firstgen"])2.4.3 Attributes of lists and data frames
Attributes of a named list:
list2 <- list(col_a = c(1,2,3), col_b = c(4,5,6))
str(list2)List of 2
$ col_a: num [1:3] 1 2 3
$ col_b: num [1:3] 4 5 6attributes(list2)$names
[1] "col_a" "col_b"Note that the names attribute is an attribute of the list, not an attribute of the elements within the list (which are atomic vectors)
list2[['col_a']] # the element named 'col_a'[1] 1 2 3str(list2[['col_a']]) # structure of the element named 'col_a' num [1:3] 1 2 3attributes(list2[['col_a']]) # attributes of element named 'col_a'NULLAttributes of a data frame:
list3 <- data.frame(col_a = c(1,2,3), col_b = c(4,5,6))
str(list3)'data.frame': 3 obs. of 2 variables:
$ col_a: num 1 2 3
$ col_b: num 4 5 6attributes(list3)$names
[1] "col_a" "col_b"
$class
[1] "data.frame"
$row.names
[1] 1 2 3Note: attributes names, class and row.names are attributes of the data frame
- they are not attributes of the elements (variables) within the data frame, which are atomic vectors (i.e., just the data)
str(list3[['col_a']]) # structure of the element named 'col_a' num [1:3] 1 2 3attributes(list3[['col_a']]) # attributes of element named 'col_a'NULL2.4.4 attr() function to get or set specific attributes of an object
Syntax
- Get:
attr(x, which, exact = FALSE) - Set:
attr(x, which) <- value
Arguments
x: an object whose attributes are to be accessedwhich: a non-empty character string specifying which attribute is to be accessedexact(logical): shouldwhichbe matched exactly? default isexact = FALSEvalue: an object, new value of attribute, orNULLto remove attribute
Using attr() to get specific attribute of an object
vector1 <- c(a = 1, b= 2, c= 3, d = 4)
attributes(vector1)$names
[1] "a" "b" "c" "d"attr(x=vector1, which = "names", exact = FALSE)[1] "a" "b" "c" "d"attr(vector1, "names")[1] "a" "b" "c" "d"attr(vector1, "name") # we don't provide exact name of attribute[1] "a" "b" "c" "d"attr(vector1, "name", exact = TRUE) # don't provide exact name of attributeNULLUsing attr() to set specific attribute of an object (output omitted)
(vector1 <- c(a = 1, b= 2, c= 3, d = 4))
attributes(vector1) # see all attributes
attr(x=vector1, which = "greeting") <- "Hi!" # create new attribute
attr(x=vector1, which = "greeting") # see attribute
attr(vector1, "farewell") <- "Bye!" # create attribute
attr(x=vector1, which = "names") # see names attribute
attr(x=vector1, which = "names") <- NULL # delete names attribute
attributes(vector1) # see all attributes2.4.5 attr() function on data frames
Using wwlist, create data frame with three variables
wwlist_small <- wwlist[1:25, ] %>% select(hs_state,firstgen,med_inc_zip)
str(wwlist_small)
attributes(wwlist_small)
attributes(wwlist_small) %>% str()Get/set attribute of a data frame
#get/examine names attribute
attr(x=wwlist_small, which = "names")
str(attr(x=wwlist_small, which = "names")) # names attribute is character atomic vector, length=3
#add new attribute to data frame
attr(x=wwlist_small, which = "new_attribute") <- "contents of new attribute"
attributes(wwlist_small)Get/set attribute of a variable in data frame
str(wwlist_small$med_inc_zip)
attributes(wwlist_small$med_inc_zip)
#create attribute for variable med_inc_zip
attr(wwlist_small$med_inc_zip, "inc attribute") <- "inc attribute contents"
#investigate attribute for variable med_inc_zip
attributes(wwlist_small$med_inc_zip)
str(wwlist_small$med_inc_zip)
attr(wwlist_small$med_inc_zip, "inc attribute")2.4.6 Why add attributes to data frame or variables of data frame?
Pedagogical reasons
- Important to know how you can apply
attributes()andattr()to data frames and to variables within data frames
Example practical application: interactive dashboards
- When creating “dashboard” you might want to add “tooltips”
- “Tooltip” is a message that appears when cursor is positioned over an icon
- The text in the tooltip is the contents of an attribute
- Example dashboard: LINK
2.4.7 Student exercise
Using
wwlist, create data frame of 30 observations with three variables:state,zip5,pop_total_zipReturn all attributes of this new data frame using
attributes(). Then, get thenamesattribute of the data frame usingattr().Add a new attribute to the data frame called
attribute_datawhose content is"new attribute of data". Then, return all attributes of the data frame as well as get the value of the newly createdattribute_data.Return the attributes of the variable
pop_total_zipin the data frame.Add a new attribute to the variable
pop_total_zipcalledattribute_variablewhose content is"new attribute of variable". Then, return all attributes of the variable as well as get the value of the newly createdattribute_variable.
Solutions to student exercise
# Part 1
wwlist_exercise <- wwlist[1:30, ] %>% select(state, zip5, pop_total_zip)
# Part 2
attributes(wwlist_exercise)
attr(x=wwlist_exercise, which = "names")
# Part 3
attr(x=wwlist_exercise, which = "attribute_data") <- "new attribute of data"
attributes(wwlist_exercise)
attr(wwlist_exercise, which ="attribute_data")
# Part 4
attributes(wwlist_exercise$pop_total_zip)
# Part 5
attr(wwlist_exercise$pop_total_zip, "attribute_variable") <- "new attribute of variable"
attributes(wwlist_exercise$pop_total_zip)
attr(wwlist_exercise$pop_total_zip, "attribute_variable")3 Object class
Every object in R has a class
- Class is an attribute of an object
- Object class controls how functions work and defines the rules for how objects can be treated by object oriented programming language
- E.g., which functions you can apply to object of a particular class
- E.g., what the function does to one object class, what it does to another object class
You can use the class() function to identify object class:
(vector2 <- c(a = 1, b= 2, c= 3, d = 4))a b c d
1 2 3 4 typeof(vector2)[1] "double"class(vector2)[1] "numeric"When I encounter a new object I often investigate object by applying typeof(), class(), and attributes() functions:
typeof(vector2)[1] "double"class(vector2)[1] "numeric"attributes(vector2)$names
[1] "a" "b" "c" "d"3.1 Why is object class important?
Specific functions usually work with only particular classes of objects
- “Date” functions usually only work on objects with a date class
- “String” functions usually only work on objects with a character class
- Functions that do mathematical computation usually work on objects with a numeric class
3.1.1 Class and object-oriented programming
R is an object-oriented programming language
Definition of object oriented programming from this LINK
“Object-oriented programming (OOP) refers to a type of computer programming in which programmers define not only the data type of a data structure, but also the types of operations (functions) that can be applied to the data structure.”
Object class is fundamental to object oriented programming because:
- Object class determines which functions can be applied to the object
- Object class also determines what those functions do to the object
- E.g., a specific function might do one thing to objects of class A and another thing to objects of class B
- What a function does to objects of different class is determined by whoever wrote the function
Many different object classes exist in R
- You can also create your own classes
- Example: the
labelledclass is an object class created by Hadley Wickham when he created thehavenpackage
- Example: the
- In this course we will work with classes that have been created by others
3.1.2 Functions care about object class, not object type
Example: sum() applies to numeric, logical, or complex class objects
Apply sum() to object with class = logical:
x <- c(TRUE, FALSE, NA, TRUE)
typeof(x)[1] "logical"class(x)[1] "logical"sum(x, na.rm = TRUE)[1] 2Apply sum() to object with class = numeric:
typeof(wwlist$med_inc_zip) [1] "double"class(wwlist$med_inc_zip) [1] "numeric"wwlist$med_inc_zip[1:5][1] 92320.5 63653.0 88344.5 88408.5 82895.0sum(wwlist$med_inc_zip[1:5], na.rm = TRUE) [1] 415621.5What happens when we try to apply sum() to an object with class = character?
typeof(wwlist$hs_city)
class(wwlist$hs_city)
wwlist$hs_city[1:5]
sum(wwlist$hs_city[1:5], na.rm = TRUE) Example: year() from lubridate package applies to date-time objects
Apply year() to object with class = Date:
wwlist$receive_date[1:5][1] "2016-05-31" "2016-05-31" "2016-05-31" "2016-05-31" "2016-05-31"typeof(wwlist$receive_date)[1] "double"class(wwlist$receive_date) [1] "Date"year(wwlist$receive_date[1:5])[1] 2016 2016 2016 2016 2016What happens when we try to apply year() to an object with class = numeric?
typeof(wwlist$med_inc_zip)
class(wwlist$med_inc_zip)
year(wwlist$med_inc_zip[1:10]) Example: tolower() applies to character class objects
- Syntax:
tolower(x) xis “a character vector, or an object that can be coerced to character byas.character()”
Most string functions are intended to apply to objects with a character class
- type = character
- class = character
Apply tolower() to object with class = character:
str(wwlist$hs_city) chr [1:268396] "Seattle" "Covington" "Everett" "Seattle" "Lake Stevens" ...typeof(wwlist$hs_city)[1] "character"class(wwlist$hs_city)[1] "character"wwlist$hs_city[1:6][1] "Seattle" "Covington" "Everett" "Seattle" "Lake Stevens"
[6] "Seattle" tolower(wwlist$hs_city[1:6])[1] "seattle" "covington" "everett" "seattle" "lake stevens"
[6] "seattle" 4 Class == factor
Recoding variable ethn_code from data frame wwlist
Let’s first recode the ethn_code variable:
wwlist <- wwlist %>%
mutate(ethn_code =
recode(ethn_code,
"american indian or alaska native" = "nativeam",
"asian or native hawaiian or other pacific islander" = "api",
"black or african american" = "black",
"cuban" = "latinx",
"mexican/mexican american" = "latinx",
"not reported" = "not_reported",
"other-2 or more" = "multirace",
"other spanish/hispanic" = "latinx",
"puerto rican" = "latinx",
"white" = "white"
)
)
str(wwlist$ethn_code)
wwlist %>% count(ethn_code)4.1 Factors
Factors are an object class used to display categorical data (e.g., marital status)
- A factor is an augmented vector built by attaching a levels attribute to an (atomic) integer vectors
Usually, we would prefer a categorical variable (e.g., race, school type) to be a factor variable rather than a character variable
- So far in the course I have made all categorical variables character variables because we have not introduced factors yet
Create factor version of character variable ethn_code using base R factor() function:
str(wwlist$ethn_code) chr [1:268396] "multirace" "white" "white" "multirace" "white" "multirace" ...class(wwlist$ethn_code)[1] "character"# create factor var; tidyverse approach
wwlist <- wwlist %>% mutate(ethn_code_fac = factor(ethn_code))
#wwlist$ethn_code_fac <- factor(wwlist$ethn_code) # base r approach
str(wwlist$ethn_code) chr [1:268396] "multirace" "white" "white" "multirace" "white" "multirace" ...str(wwlist$ethn_code_fac) Factor w/ 7 levels "api","black",..: 4 7 7 4 7 4 4 4 4 7 ...Character variable ethn_code:
typeof(wwlist$ethn_code)[1] "character"class(wwlist$ethn_code)[1] "character"attributes(wwlist$ethn_code)NULLstr(wwlist$ethn_code) chr [1:268396] "multirace" "white" "white" "multirace" "white" "multirace" ...Factor variable ethn_code_fac:
typeof(wwlist$ethn_code_fac)[1] "integer"class(wwlist$ethn_code_fac)[1] "factor"attributes(wwlist$ethn_code_fac)$levels
[1] "api" "black" "latinx" "multirace" "nativeam"
[6] "not_reported" "white"
$class
[1] "factor"str(wwlist$ethn_code_fac) Factor w/ 7 levels "api","black",..: 4 7 7 4 7 4 4 4 4 7 ...4.2 Working with factor variables
Main things to note about variable ethn_code_fac
- type = integer
- class = factor, because the variable has a levels attribute
- Underlying data are integers, but the values of the levels attribute is what’s displayed:
# Print first few obs of ethn_code_fac
wwlist$ethn_code_fac[1:5][1] multirace white white multirace white
Levels: api black latinx multirace nativeam not_reported white# Print count for each category in ethn_code_fac
wwlist %>% count(ethn_code_fac)# A tibble: 7 × 2
ethn_code_fac n
<fct> <int>
1 api 2385
2 black 563
3 latinx 9245
4 multirace 90584
5 nativeam 202
6 not_reported 5737
7 white 159680Apply as.integer() to display underlying integer values of factor the variable
Investigate as.integer() function:
typeof(wwlist$ethn_code_fac)[1] "integer"class(wwlist$ethn_code_fac)[1] "factor"typeof(as.integer(wwlist$ethn_code_fac))[1] "integer"class(as.integer(wwlist$ethn_code_fac))[1] "integer"Display underlying integer values of variable ethn_code_fac:
wwlist %>% count(as.integer(ethn_code_fac))# A tibble: 7 × 2
`as.integer(ethn_code_fac)` n
<int> <int>
1 1 2385
2 2 563
3 3 9245
4 4 90584
5 5 202
6 6 5737
7 7 159680Refer to categories of a factor (e.g., when filtering obs) using values of levels attribute rather than underlying values of variable
- Values of levels attribute for
ethn_code_fac(output omitted)
attributes(wwlist$ethn_code_fac)Example: Count the number of prospects in wwlist who identify as “white”
# referring to variable value; this doesn't work
wwlist %>% filter(ethn_code_fac==7) %>% count() # A tibble: 1 × 1
n
<int>
1 0#referring to value of level attribute; this works
wwlist %>% filter(ethn_code_fac=="white") %>% count()# A tibble: 1 × 1
n
<int>
1 159680Example: Count the number of prospects in wwlist who identify as “white”
- To refer to underlying integer values, apply
as.integer()function to factor variable
attributes(wwlist$ethn_code_fac)$levels
[1] "api" "black" "latinx" "multirace" "nativeam"
[6] "not_reported" "white"
$class
[1] "factor"wwlist %>% filter(as.integer(ethn_code_fac)==7) %>% count# A tibble: 1 × 1
n
<int>
1 1596804.3 How to identify the variable values associated with factor levels
Create a factor version of the character variable psat_range
wwlist %>% count(psat_range)
wwlist <- wwlist %>% mutate(psat_range_fac = factor(psat_range))
wwlist %>% count(psat_range_fac)
attributes(wwlist$psat_range_fac)Investigate values associated with factor levels using levels() and nlevels()
levels(wwlist$psat_range_fac) #starts at 1
nlevels(wwlist$psat_range_fac) #7 levels total
levels(wwlist$psat_range_fac)[1:3] #prints levels 1-3Once values associated with factor levels are known:
- Can filter based on underlying integer values using
as.integer()
wwlist %>% filter(as.integer(psat_range_fac)==4) %>% count()# A tibble: 1 × 1
n
<int>
1 8348- Or filter based on value of factor levels
wwlist %>% filter(psat_range=="1270-1520") %>% count()# A tibble: 1 × 1
n
<int>
1 83484.3.1 Student exercise working with factors
- After running the code below, use
typeof(),class(),str(), andattributes()functions to check the new variablereceive_year
- Create a factor variable from the input variable
receive_yearand name itreceive_year_fac
- Run the same functions (
typeof(),class(), etc.) from the first question using the new variable you created
- Get a count of
receive_year_fac. (hint: you could also run this in the console to see values associated with each factor)
Run this code to create a year variable from the input variable receive_date:
# wwlist %>% glimpse()
library(lubridate) # load library if you haven't already
wwlist <- wwlist %>%
mutate(receive_year = year(receive_date)) # create year variable with lubridate
# Check variable
wwlist %>%
count(receive_year)
wwlist %>%
group_by(receive_year) %>%
count(receive_date)
Solutions to student exercise working with factors %>%
- After running the code below, use
typeof(),class(),str(), andattributes()functions to check the new variablereceive_year
typeof(wwlist$receive_year)[1] "double"class(wwlist$receive_year)[1] "numeric"str(wwlist$receive_year) num [1:268396] 2016 2016 2016 2016 2016 ...attributes(wwlist$receive_year) NULL- Create a factor variable from the input variable
receive_yearand name itreceive_year_fac
# create factor var; tidyverse approach
wwlist <- wwlist %>%
mutate(receive_year_fac = factor(receive_year)) - Run the same functions (
typeof(),class(), etc.) from the first question using the new variable you created
typeof(wwlist$receive_year_fac)[1] "integer"class(wwlist$receive_year_fac)[1] "factor"str(wwlist$receive_year_fac) Factor w/ 3 levels "2016","2017",..: 1 1 1 1 1 1 1 1 1 1 ...attributes(wwlist$receive_year_fac) $levels
[1] "2016" "2017" "2018"
$class
[1] "factor"- Get a count of
receive_year_fac. (hint: you could also run this in the console to see values associated with each factor)
wwlist %>%
count(receive_year_fac)# A tibble: 3 × 2
receive_year_fac n
<fct> <int>
1 2016 89637
2 2017 89816
3 2018 889435 Class == labelled
5.1 Data we will use to introduce labelled class
High school longitudinal surveys from National Center for Education Statistics (NCES)
- Follow U.S. students from high school through college, labor market
We will be working with High School Longitudinal Study of 2009 (HSLS:09)
- Follows 9th graders from 2009
- Data collection waves
- Base Year (2009)
- First Follow-up (2012)
- 2013 Update (2013)
- High School Transcripts (2013-2014)
- Second Follow-up (2016)
5.2 Using haven package to read SAS/SPSS/Stata datasets into R
haven, which is part of tidyverse, “enables R to read and write various data formats” from the following statistical packages:
- SAS
- SPSS
- Stata
If you haven’t installed haven run the following code in your console.
- Generic syntax:
install.packages("package_name") - Install “haven”:
install.packages("haven")
Make sure to load haven (e.g., library(haven))
When using haven to read data, resulting R objects have these characteristics:
- Data frames are tibbles, Tidyverse’s preferred class of data frames
- Transform variables with “value labels” into the
labelled()classlabelledis an object class, just likefactoris an object classlabelledis an object class created by folks who createdhavenpackagelabelledandfactorclasses are both viable alternatives for categorical variables- Helpful description of
labelledclass HERE
- Dates and times converted to R date/time classes
- Character vectors not converted to factors
Use read_dta() function from haven package to import Stata dataset into R
hsls <- read_dta(file="https://github.com/ozanj/rclass/raw/master/data/hsls/hsls_stu_small.dta")Must run this code chunk; permanently changes uppercase variable names to lowercase
names(hsls)
names(hsls) <- tolower(names(hsls)) # convert names to lowercase
names(hsls) # names now lowercase
str(hsls) # ughInvestigate variable s3classes from data frame hsls
- Identifies whether respondent taking postsecondary classes as of 11/1/2013
typeof(hsls$s3classes)
class(hsls$s3classes)
str(hsls$s3classes)Investigate attributes of s3classes
attributes(hsls$s3classes) # all attributes
#specific attributes: using syntax: attr(x, which, exact = FALSE)
attr(x=hsls$s3classes, which = "label") # label attribute
attr(x=hsls$s3classes, which = "labels") # labels attribute5.3 What is object class = labelled?
Variable labels are labels attached to a specific variable (e.g., marital status) Value labels [in Stata] are labels attached to specific values of a variable, e.g.:
- Var value
1attached to value label “married”,2=“single”,3=“divorced”
labelled is object class for importing vars with value labels from SAS/SPSS/Stata
labelledobject class created byhavenpackage- Characteristics of variables in R data frame with
class==labelled:- Data
typecan be numeric(double) or character - To see
value labelsassociated with each value:attr(df_name$var_name,"labels")- E.g.,
attr(hsls$s3classes,"labels")
- Data
Investigate the attributes of hsls$s3classes
typeof(hsls$s3classes)
class(hsls$s3classes)
str(hsls$s3classes)
attributes(hsls$s3classes)Use attr(object_name,"attribute_name") to refer to each attribute
attr(hsls$s3classes,"label")
attr(hsls$s3classes,"format.stata")
attr(hsls$s3classes,"class")
attr(hsls$s3classes,"labels")5.4 labelled package
Purpose of the labelled package is to work with data imported from SPSS/Stata/SAS using the haven package
labelledpackage contains functions to work with objects that havelabelledclass- From package documentation:
- “purpose of the
labelledpackage is to provide functions to manipulate metadata as variable labels, value labels and defined missing values using thelabelledclass and thelabelattribute introduced inhavenpackage.”
- “purpose of the
- More info on the
labelledpackage: LINK
Functions in labelled package
5.5 Get variable and value labels
Get variable labels using var_label()
hsls %>% select(s3classes) %>% var_label()$s3classes
[1] "S3 B01A Taking postsecondary classes as of Nov 1 2013"Get value labels using val_labels()
hsls %>% select(s3classes) %>% val_labels()$s3classes
Missing
-9
Unit non-response
-8
Item legitimate skip/NA
-7
Component not applicable
-6
Item not administered: abbreviated interview
-4
Yes
1
No
2
Don't know
3 5.6 Working with labelled class data
Create frequency tables with labelled class variables using count()
- when running individual code chunks, default setting is to show variable values not value labels
hsls %>% count(s3classes)# A tibble: 5 × 2
s3classes n
<dbl+lbl> <int>
1 -9 [Missing] 59
2 -8 [Unit non-response] 4945
3 1 [Yes] 13477
4 2 [No] 3401
5 3 [Don't know] 1621when running individual code chunks, to make frequency table show value labels add %>% as_factor() to pipe
as_factor()is function fromhaventhat converts an object to a factor
hsls %>% count(s3classes) %>% as_factor()# A tibble: 5 × 2
s3classes n
<fct> <int>
1 Missing 59
2 Unit non-response 4945
3 Yes 13477
4 No 3401
5 Don't know 1621To isolate values of labelled class variables in filter() function:
- Refer to variable value, not the value label
Task
- How many observations in var
s3classesassociated with “Unit non-response” - How many observations in var
s3classesassociated with “Yes”
General steps to follow:
- Investigate object
- Use
filter()to isolate desired observations
Investigate object
class(hsls$s3classes)
hsls %>% select(s3classes) %>% var_label() #show variable label
hsls %>% select(s3classes) %>% val_labels() #show value label
hsls %>% count(s3classes) # freq table, values
hsls %>% count(s3classes) %>% as_factor() # freq table, value labelsFilter specific values
hsls %>% filter(s3classes==-8) %>% count() # -8 = unit non-response
hsls %>% filter(s3classes==1) %>% count() # 1 = yes5.7 Set variable and value labels
Set variable labels using var_label() or set_variable_labels()
# Set one variable label
var_label(df_name$var_name) <- 'variable label'
# Set multiple variable labels
df_name <- df_name %>%
set_variable_labels(
var_name_1 = 'variable label 1',
var_name_2 = 'variable label 2',
var_name_3 = 'variable label 3'
)Set value labels using val_label() or set_value_labels()
# Set one value label
val_label(df_name$var_name, 'variable_value') <- 'value_label'
# Set multiple value labels
df_name <- df_name %>%
set_value_labels(
var_name_1 = c('value_label_1' = 'variable_value_1',
'value_label_2' = 'variable_value_2',
var_name_2 = c('value_label_3' = 'variable_value_3',
'value_label_4' = 'variable_value_4')
)5.7.1 Create example data frame
df <- tribble(
~id, ~edu, ~sch,
#--|--|----
1, 2, 2,
2, 1, 1,
3, 3, 2,
4, 4, 2,
5, 1, 2
)
df# A tibble: 5 × 3
id edu sch
<dbl> <dbl> <dbl>
1 1 2 2
2 2 1 1
3 3 3 2
4 4 4 2
5 5 1 2str(df)tibble [5 × 3] (S3: tbl_df/tbl/data.frame)
$ id : num [1:5] 1 2 3 4 5
$ edu: num [1:5] 2 1 3 4 1
$ sch: num [1:5] 2 1 2 2 25.7.2 Set variable labels
Use set_variable_labels() or var_label() to manually set variable labels
str(df$sch) num [1:5] 2 1 2 2 2var_label(df$sch)NULL# Using set_variable_labels()
df <- df %>%
set_variable_labels(
id = "Unique identification number",
edu = "Education level"
)
# Using var_label()
var_label(df$sch) <- 'Type of school attending'
str(df$sch) num [1:5] 2 1 2 2 2
- attr(*, "label")= chr "Type of school attending"var_label(df$sch)[1] "Type of school attending"5.7.3 Set value labels
Use set_value_labels() or val_label() to manually set value labels
val_labels(df$sch)NULL# Using set_value_labels()
df <- df %>%
set_value_labels(
edu = c('High School' = 1,
'AA degree' = 2,
'BA degree' = 3,
'MA or higher' = 4),
sch = c('Private' = 1))
# Using val_label()
val_label(df$sch, 2) <- 'Public'
str(df$sch) dbl+lbl [1:5] 2, 1, 2, 2, 2
@ labels: Named num [1:2] 1 2
..- attr(*, "names")= chr [1:2] "Private" "Public"
@ label : chr "Type of school attending"val_labels(df$sch)Private Public
1 2 5.7.4 View the set variable and value labels
# View variable and value labels using attributes()
attributes(df$sch)$labels
Private Public
1 2
$label
[1] "Type of school attending"
$class
[1] "haven_labelled" "vctrs_vctr" "double" # View variable label
var_label(df$sch)[1] "Type of school attending"attr(df$sch, 'label')[1] "Type of school attending"# View value labels
val_labels(df$sch)Private Public
1 2 attr(df$sch, 'labels')Private Public
1 2 5.7.5 labelled student exercise
- Get variable and value labels of the variable
s3hsin thehslsdata frame - Get a count of the variable
s3hsshowing the values and the value labels (hint: useas_factor()) - Get a count of the rows whose value for
s3hsis associated with “Missing” (hint: usefilter()) - Get a count of the rows whose value for
s3hsis associated with “Missing” or “Unit non-response” - Add variable label for
pop_asian_zip&pop_asian_statein data framewwlist - Add value labels for
ethn_codein data framewwlist
labelled student exercise solutions
- Get variable and value labels of the variable
s3hsin thehslsdata frame
hsls %>%
select(s3hs) %>%
var_label() $s3hs
[1] "S3 B01F Attending high school or homeschool as of Nov 1 2013"hsls %>%
select(s3hs) %>%
val_labels()$s3hs
Missing
-9
Unit non-response
-8
Item legitimate skip/NA
-7
Component not applicable
-6
Item not administered: abbreviated interview
-4
Yes
1
No
2
Don't know
3 - Get a count of the variable
s3hsshowing the values and the value labels (hint: useas_factor())
hsls %>%
count(s3hs) # A tibble: 6 × 2
s3hs n
<dbl+lbl> <int>
1 -9 [Missing] 22
2 -8 [Unit non-response] 4945
3 -7 [Item legitimate skip/NA] 16770
4 1 [Yes] 624
5 2 [No] 985
6 3 [Don't know] 157hsls %>%
count(s3hs) %>%
as_factor() # A tibble: 6 × 2
s3hs n
<fct> <int>
1 Missing 22
2 Unit non-response 4945
3 Item legitimate skip/NA 16770
4 Yes 624
5 No 985
6 Don't know 157- Get a count of the rows whose value for
s3hsis associated with “Missing” (hint: usefilter())
hsls %>%
filter(s3hs== -9) %>%
count()# A tibble: 1 × 1
n
<int>
1 22- Get a count of the rows whose value for
s3hsis associated with “Missing” or “Unit non-response”
hsls %>%
filter(s3hs== -9 | s3hs== -8) %>%
count()# A tibble: 1 × 1
n
<int>
1 4967- Add variable label for
pop_asian_zip&pop_asian_statein data framewwlist
# variable labels
wwlist %>% select(pop_asian_zip, pop_asian_state) %>% var_label()$pop_asian_zip
NULL
$pop_asian_state
NULL# set variable labels
wwlist <- wwlist %>%
set_variable_labels(
pop_asian_zip = "total asian population in zip",
pop_asian_state ="total asian population in state"
)
# attribute of variable
attributes(wwlist$pop_asian_zip)$label
[1] "total asian population in zip"attributes(wwlist$pop_asian_state)$label
[1] "total asian population in state"- Add value labels for
ethn_codein data framewwlist
# count
wwlist %>% count(ethn_code)
# value labels
wwlist %>% select(ethn_code) %>% val_labels
# set value labels to ethn_code variable
wwlist <- wwlist %>%
set_value_labels(
ethn_code = c("asian or native hawaiian or other pacific islander" = "api",
"black or african american" = "black",
"cuban or mexican/mexican american or other spanish/hispanic or puerto rican" = "latinx",
"other-2 or more" = "multirace",
"american indian or alaska native" = "nativeam",
"not reported" = "not_reported",
"white" = "white"
)
)6 Comparing class labelled to class factor
class==labelled |
class==factor |
|
|---|---|---|
| data type | numeric or character | integer |
| name of value label attribute | labels | levels |
| refer to data using | variable values | levels attribute |
So should you work with class==labelled or class==factor?
- No right or wrong answer; this is a subjective decision
- Personally, I prefer
factorclass- Easier to run analysis (see section below)
labelledclass- Feels more suited to working with survey data variables, where there are usually several different values that represent different kinds of “missing” values.
- You can use both! We recommend using labelled variables, particularly for exploratory data analysis, and factor variables for analysis when using categorical variables.
6.0.1 Why factor variables are essential for analysis
When running analyses using categorical variables (e.g., marital status: single, married, divorced, etc.), it is best practice to use class == factor over class == labelled.
To demonstrate a quick example, let’s load data from ELS. We will use this data set later in the graphing and ggplot lecture, so don’t worry about understanding it now.
Say we are interested in the relationship between having an internship or co-op job (X) (in 2005-2006) on subsequent earnings (Y) (in 2011) after controlling for standardized reading test score (as a measure for high school achievement), sex, and race/ethnicity.
- We have two linear models below, the first using variables that are class labelled and the second using variables that are class factor.
Linear model lm() model using categorical variables of class labelled
$label
[1] "F1 sex-composite"
$format.stata
[1] "%8.0g"
$class
[1] "haven_labelled" "vctrs_vctr" "double"
$labels
Male Female
1 2 $labels
white api black latinx native multi
1 2 3 4 5 6
$class
[1] "haven_labelled" "vctrs_vctr" "double"
$label
[1] "categorical measure of race based on variable f1race"intern_mod1 <- lm(formula = f3ern2011 ~ f2intern0506 + bytxrstd + f1sex + f1race_v2, data = df_els_stu_allobs %>% filter(f2enroll0506==1))
summary(intern_mod1)
Call:
lm(formula = f3ern2011 ~ f2intern0506 + bytxrstd + f1sex + f1race_v2,
data = df_els_stu_allobs %>% filter(f2enroll0506 == 1))
Residuals:
Min 1Q Median 3Q Max
-37135 -18595 -2783 12020 231894
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 20479.35 1607.58 12.739 < 2e-16 ***
f2intern0506 5580.01 1059.32 5.268 1.41e-07 ***
bytxrstd 261.16 23.86 10.945 < 2e-16 ***
f1sex -4104.21 515.28 -7.965 1.85e-15 ***
f1race_v2 -1275.57 189.17 -6.743 1.64e-11 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 24760 on 9317 degrees of freedom
(21 observations deleted due to missingness)
Multiple R-squared: 0.02949, Adjusted R-squared: 0.02908
F-statistic: 70.78 on 4 and 9317 DF, p-value: < 2.2e-16Linear model lm() model using categorical variables of class factor
$levels
[1] "Male" "Female"
$class
[1] "factor"
$label
[1] "F1 sex-composite"$levels
[1] "white" "api" "black" "latinx" "native" "multi"
$class
[1] "factor"
$label
[1] "categorical measure of race based on variable f1race"intern_mod2 <- lm(formula = f3ern2011 ~ f2intern0506 + bytxrstd + f1sex + f1race_v2, data = df_els_stu_allobs_fac %>% filter(f2enroll0506=="yes"))
summary(intern_mod2)
Call:
lm(formula = f3ern2011 ~ f2intern0506 + bytxrstd + f1sex + f1race_v2,
data = df_els_stu_allobs_fac %>% filter(f2enroll0506 == "yes"))
Residuals:
Min 1Q Median 3Q Max
-37282 -18246 -2868 12045 228779
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 16503.51 1414.69 11.666 < 2e-16 ***
f2intern0506yes 5616.95 1059.04 5.304 1.16e-07 ***
bytxrstd 240.60 24.38 9.869 < 2e-16 ***
f1sexFemale -4055.06 515.10 -7.872 3.87e-15 ***
f1race_v2api -1784.73 817.78 -2.182 0.02910 *
f1race_v2black -5113.41 859.37 -5.950 2.78e-09 ***
f1race_v2latinx -4768.47 838.71 -5.685 1.34e-08 ***
f1race_v2native -10149.40 3591.46 -2.826 0.00472 **
f1race_v2multi -3206.89 1279.05 -2.507 0.01218 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 24740 on 9313 degrees of freedom
(21 observations deleted due to missingness)
Multiple R-squared: 0.03148, Adjusted R-squared: 0.03065
F-statistic: 37.84 on 8 and 9313 DF, p-value: < 2.2e-16Why does this matter?
Notice that for the linear model intern_mod1, the coefficients for categorical variables sex and race are grouped together, whereas in the second linear model intern_mod2, using class factor variables, coefficients are calculated separately by “category” (e.g., female, male, black, white, Asian, etc.).
6.1 Converting class==labelled to class==factor
The as_factor() function from haven package converts variables with class==labelled to class==factor
- Can be used for descriptive statistics
hsls %>% select(s3classes) %>% count(s3classes)
hsls %>% select(s3classes) %>% count(s3classes) %>% as_factor()- Can create object with some or all
labelledvars converted tofactor
hsls_f <- as_factor(hsls, only_labelled = TRUE)Let’s examine this object
glimpse(hsls_f)
hsls_f %>% select(s3classes,s3clglvl) %>% str()
typeof(hsls_f$s3classes)
class(hsls_f$s3classes)
attributes(hsls_f$s3classes)
hsls_f %>% select(s3classes) %>% var_label()
hsls_f %>% select(s3classes) %>% val_labels()6.2 Working with class==factor data
Showing factor levels associated with a factor variable
hsls_f %>% count(s3classes)# A tibble: 5 × 2
s3classes n
<fct> <int>
1 Missing 59
2 Unit non-response 4945
3 Yes 13477
4 No 3401
5 Don't know 1621Showing variable values associated with a factor variable
hsls_f %>% count(as.integer(s3classes))# A tibble: 5 × 2
`as.integer(s3classes)` n
<int> <int>
1 1 59
2 2 4945
3 6 13477
4 7 3401
5 8 1621When sub-setting observations (e.g., filtering), refer to the level attribute, not the underlying variable integer value.
hsls_f %>% filter(s3classes=="Yes") %>% count(s3classes)# A tibble: 1 × 2
s3classes n
<fct> <int>
1 Yes 134777 Appendix: Creating factor variables
7.1 Create factors [from string variables]
To create a factor variable from string variable:
- Create a character vector containing underlying data
- Create a vector containing valid levels
- Attach levels to the data using the
factor()function
# Underlying data: months my fam is born
x1 <- c("Jan", "Aug", "Apr", "Mar")
# Create vector with valid levels
month_levels <- c("Jan", "Feb", "Mar", "Apr", "May", "Jun",
"Jul", "Aug", "Sep", "Oct", "Nov", "Dec")
# Attach levels to data
x2 <- factor(x1, levels = month_levels)Note how attributes differ:
str(x1) chr [1:4] "Jan" "Aug" "Apr" "Mar"str(x2) Factor w/ 12 levels "Jan","Feb","Mar",..: 1 8 4 3Sorting also differs:
sort(x1)[1] "Apr" "Aug" "Jan" "Mar"sort(x2)[1] Jan Mar Apr Aug
Levels: Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov DecLet’s create a character version of variable hs_state and then turn it into a factor:
#wwlist %>%
# count(hs_state)
# Subset obs to West Coast states
wwlist_temp <- wwlist %>%
filter(hs_state %in% c("CA", "OR", "WA"))
# Create character version of high school state for West Coast states only
wwlist_temp$hs_state_char <- as.character(wwlist_temp$hs_state)
# Investigate character variable
str(wwlist_temp$hs_state_char)
table(wwlist_temp$hs_state_char)
# Create new variable that assigns levels
wwlist_temp$hs_state_fac <- factor(wwlist_temp$hs_state_char, levels = c("CA","OR","WA"))
str(wwlist_temp$hs_state_fac)
attributes(wwlist_temp$hs_state_fac)
#wwlist_temp %>%
# count(hs_state_fac)
rm(wwlist_temp)How the levels argument works when underlying data is character:
- Matches value of underlying data to value of the level attribute
- Converts underlying data to integer, with level attribute attached
See Chapter 15 of Wickham for more on factors (e.g., modifying factor order, modifying factor levels)
7.2 Creating factors [from integer vectors]
Factors are just integer vectors with level attributes attached to them. So, to create a factor:
- Create a vector for the underlying data
- Create a vector that has level attributes
- Attach levels to the data using the
factor()function
a1 <- c(1,1,1,0,1,1,0) # A vector of data
a2 <- c("zero","one") # A vector of labels
# Attach labels to values
a3 <- factor(a1, labels = a2)
a3[1] one one one zero one one zero
Levels: zero onestr(a3) Factor w/ 2 levels "zero","one": 2 2 2 1 2 2 1Note: By default, factor() function attached “zero” to the lowest value of vector a1 because “zero” was the first element of vector a2
Let’s turn an integer variable into a factor variable in the wwlist data frame
Create integer version of receive_year:
#typeof(wwlist_temp$receive_year)
wwlist$receive_year_int <- as.integer(wwlist$receive_year)
str(wwlist$receive_year_int) int [1:268396] 2016 2016 2016 2016 2016 2016 2016 2016 2016 2016 ...typeof(wwlist$receive_year_int)[1] "integer"Assign levels to values of integer variable:
wwlist$receive_year_fac <- factor(wwlist$receive_year_int,
labels=c("Twenty-sixteen","Twenty-seventeen","Twenty-eighteen"))
str(wwlist$receive_year_fac)
str(wwlist$receive_year)
#Check variable
wwlist %>%
count(receive_year_fac)
wwlist %>%
count(receive_year)